Details of the Target
General Information of Target
| Target ID | LDTP04140 | |||||
|---|---|---|---|---|---|---|
| Target Name | Large ribosomal subunit protein eL29 (RPL29) | |||||
| Gene Name | RPL29 | |||||
| Gene ID | 6159 | |||||
| Synonyms |
Large ribosomal subunit protein eL29; 60S ribosomal protein L29; Cell surface heparin-binding protein HIP |
|||||
| 3D Structure | ||||||
| Sequence |
MAKSKNHTTHNQSRKWHRNGIKKPRSQRYESLKGVDPKFLRNMRFAKKHNKKGLKKMQAN
NAKAMSARAEAIKALVKPKEVKPKIPKGVSRKLDRLAYIAHPKLGKRARARIAKGLRLCR PKAKAKAKAKDQTKAQAAAPASVPAQAPKRTQAPTKASE |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Eukaryotic ribosomal protein eL29 family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function | Component of the large ribosomal subunit. The ribosome is a large ribonucleoprotein complex responsible for the synthesis of proteins in the cell. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
14.32 | LDD0402 | [1] | |
|
P3 Probe Info |
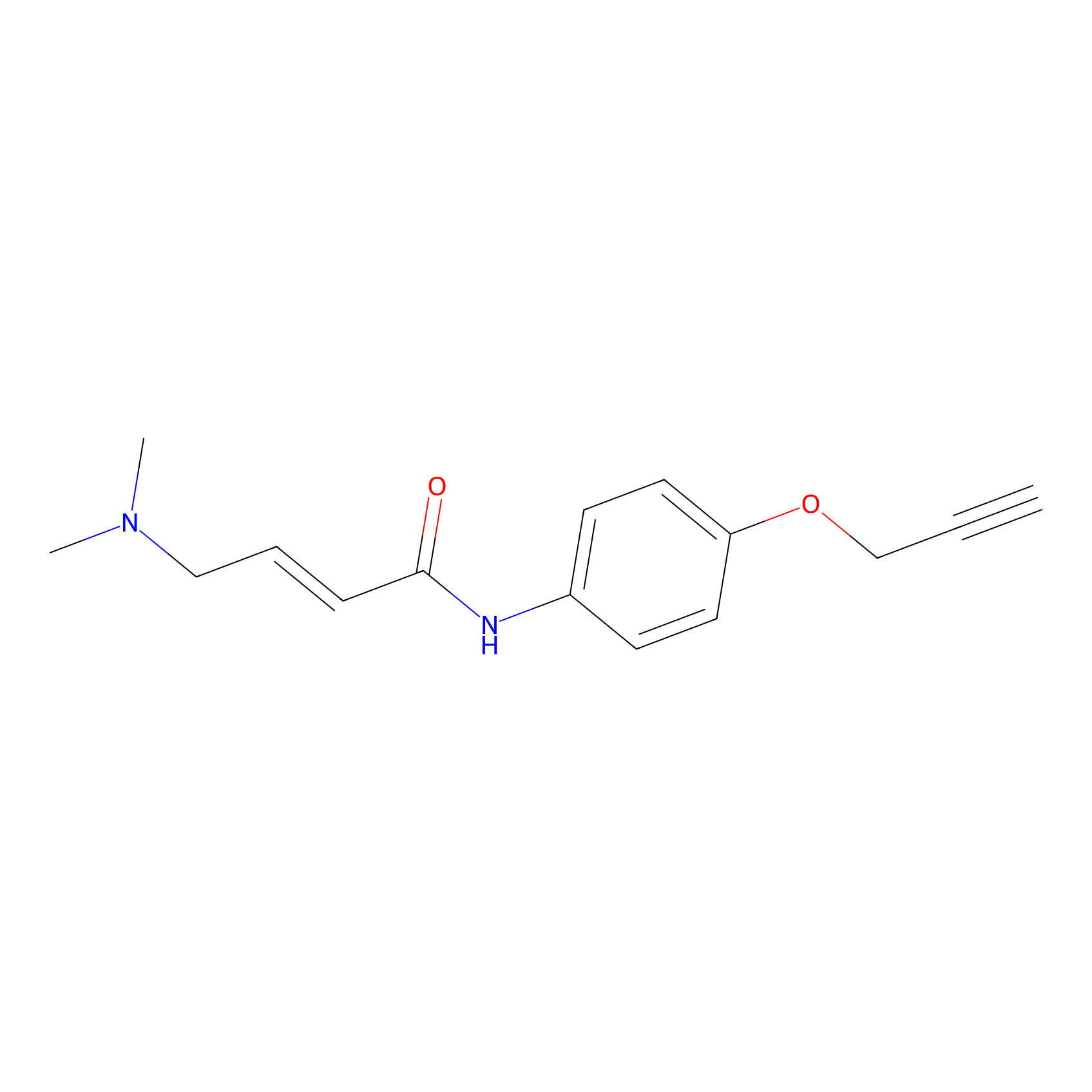 |
1.64 | LDD0450 | [2] | |
|
TH211 Probe Info |
 |
Y98(10.28) | LDD0260 | [3] | |
|
TH216 Probe Info |
 |
Y98(20.00) | LDD0259 | [3] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [4] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [4] | |
|
ONAyne Probe Info |
 |
K33(0.00); K38(0.00); K149(0.00); K156(0.00) | LDD0273 | [5] | |
|
Probe 1 Probe Info |
 |
Y98(6.10) | LDD3495 | [6] | |
|
HHS-482 Probe Info |
 |
Y98(1.04) | LDD0285 | [7] | |
|
HHS-475 Probe Info |
 |
Y98(0.78) | LDD0264 | [8] | |
|
IA-alkyne Probe Info |
 |
C119(0.60) | LDD2182 | [9] | |
|
HHS-465 Probe Info |
 |
Y98(10.00) | LDD2237 | [10] | |
|
AMP probe Probe Info |
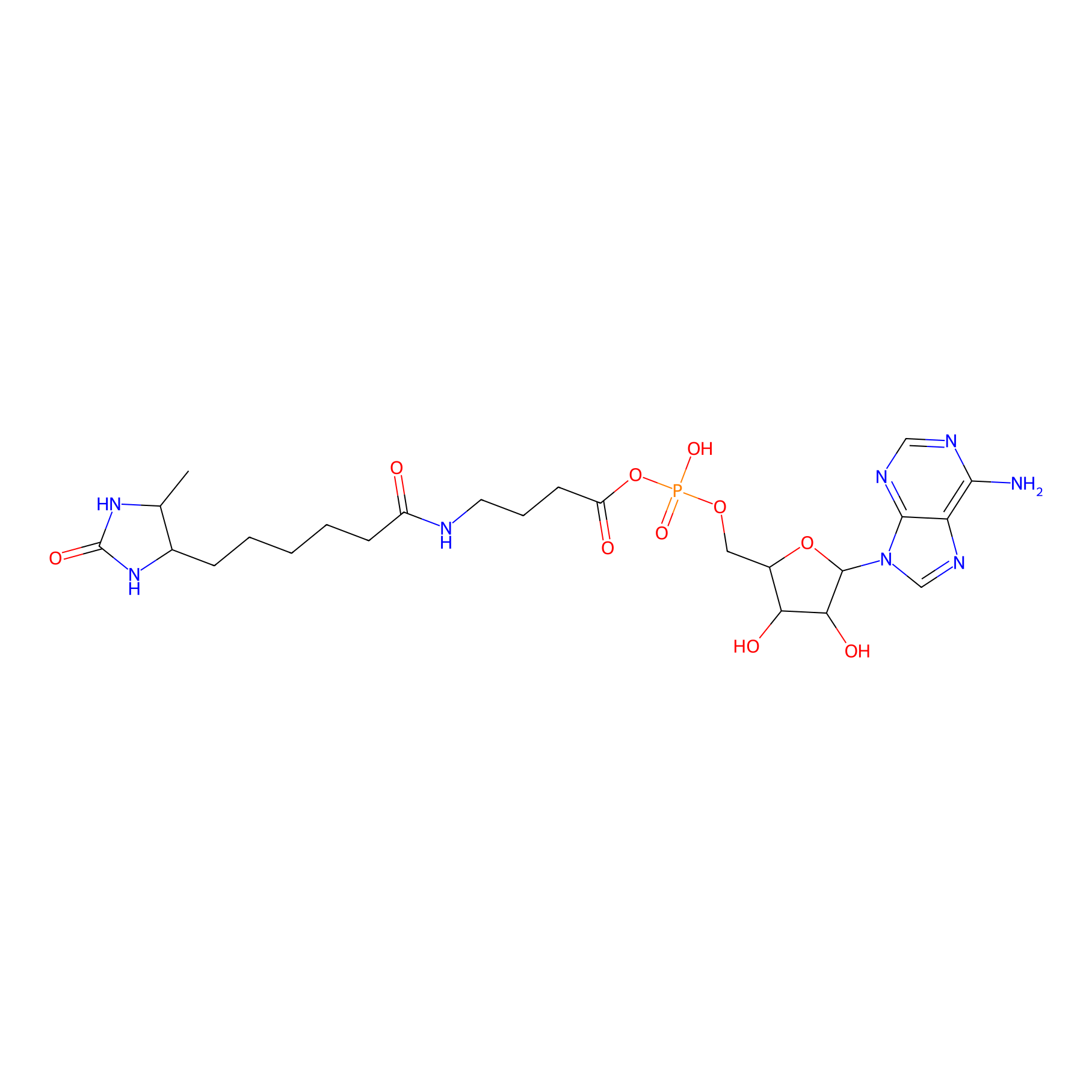 |
N.A. | LDD0200 | [11] | |
|
ATP probe Probe Info |
 |
K149(0.00); K33(0.00); K82(0.00); K84(0.00) | LDD0199 | [11] | |
|
2PCA Probe Info |
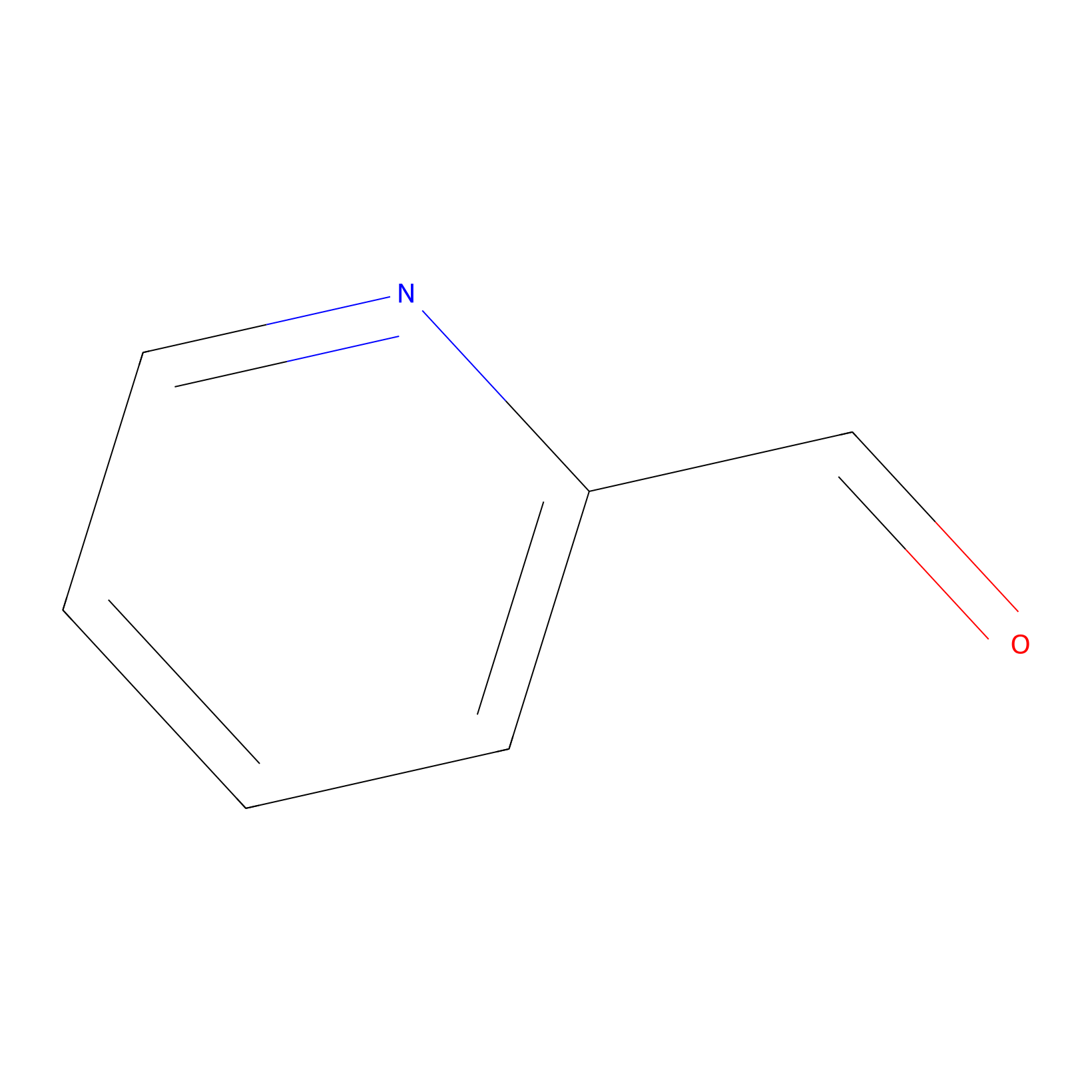 |
N.A. | LDD0034 | [12] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [13] | |
|
NHS Probe Info |
 |
K134(0.00); K73(0.00); K63(0.00); K149(0.00) | LDD0010 | [14] | |
|
OSF Probe Info |
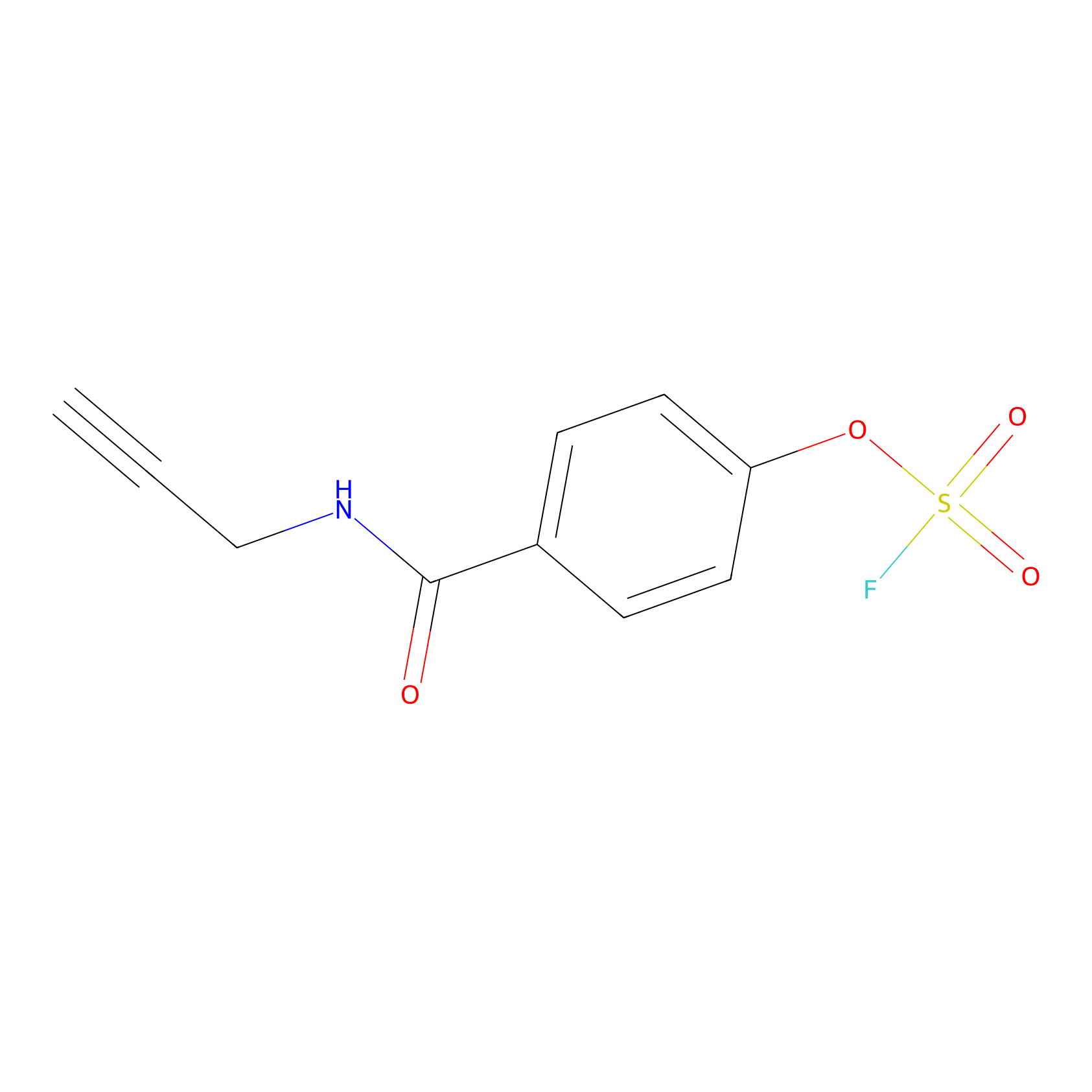 |
N.A. | LDD0029 | [15] | |
|
SF Probe Info |
 |
K149(0.00); Y29(0.00); Y98(0.00); K103(0.00) | LDD0028 | [15] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [14] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [16] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [17] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [18] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [18] | |
|
AOyne Probe Info |
 |
8.30 | LDD0443 | [19] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [20] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C187 Probe Info |
 |
17.75 | LDD1865 | [21] | |
|
C191 Probe Info |
 |
10.85 | LDD1868 | [21] | |
|
C193 Probe Info |
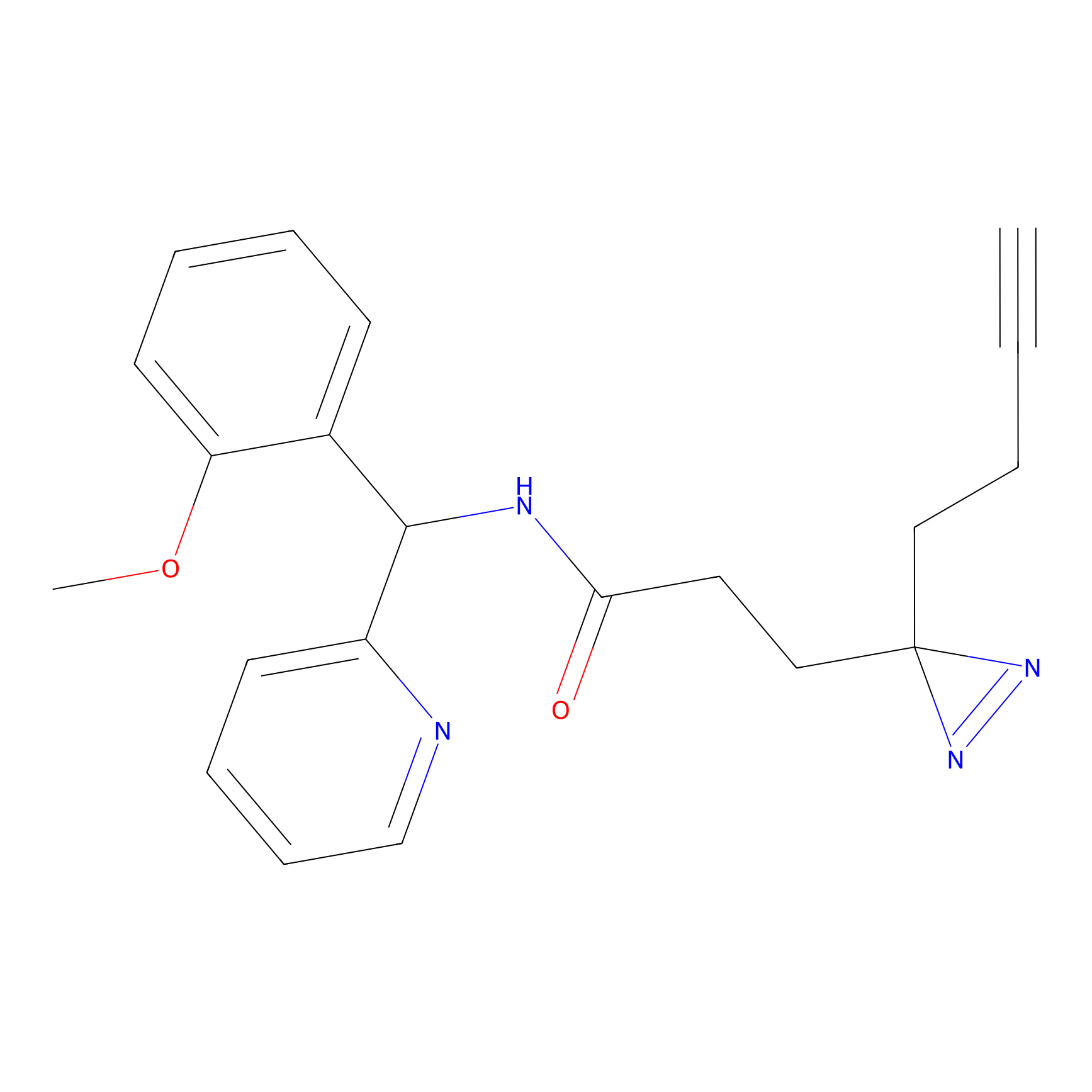 |
6.19 | LDD1869 | [21] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [22] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0156 | Aniline | NCI-H1299 | 12.92 | LDD0403 | [1] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [18] |
| LDCM0625 | F8 | Ramos | C119(5.46) | LDD2187 | [9] |
| LDCM0573 | Fragment11 | Ramos | C119(0.07) | LDD2190 | [9] |
| LDCM0576 | Fragment14 | Ramos | C119(0.29) | LDD2193 | [9] |
| LDCM0586 | Fragment28 | Ramos | C119(1.11) | LDD2198 | [9] |
| LDCM0566 | Fragment4 | Ramos | C119(0.68) | LDD2184 | [9] |
| LDCM0569 | Fragment7 | Ramos | C119(0.42) | LDD2186 | [9] |
| LDCM0116 | HHS-0101 | DM93 | Y98(0.78) | LDD0264 | [8] |
| LDCM0117 | HHS-0201 | DM93 | Y98(1.79) | LDD0265 | [8] |
| LDCM0118 | HHS-0301 | DM93 | Y98(0.61) | LDD0266 | [8] |
| LDCM0119 | HHS-0401 | DM93 | Y98(2.26) | LDD0267 | [8] |
| LDCM0120 | HHS-0701 | DM93 | Y98(1.98) | LDD0268 | [8] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [18] |
| LDCM0123 | JWB131 | DM93 | Y98(1.04) | LDD0285 | [7] |
| LDCM0124 | JWB142 | DM93 | Y98(0.86) | LDD0286 | [7] |
| LDCM0125 | JWB146 | DM93 | Y98(1.04) | LDD0287 | [7] |
| LDCM0126 | JWB150 | DM93 | Y98(2.98) | LDD0288 | [7] |
| LDCM0127 | JWB152 | DM93 | Y98(2.21) | LDD0289 | [7] |
| LDCM0128 | JWB198 | DM93 | Y98(1.32) | LDD0290 | [7] |
| LDCM0129 | JWB202 | DM93 | Y98(0.52) | LDD0291 | [7] |
| LDCM0130 | JWB211 | DM93 | Y98(0.63) | LDD0292 | [7] |
| LDCM0022 | KB02 | Ramos | C119(0.60) | LDD2182 | [9] |
| LDCM0023 | KB03 | Ramos | C119(0.57) | LDD2183 | [9] |
| LDCM0024 | KB05 | Ramos | C119(0.27) | LDD2185 | [9] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [18] |
References
