Details of the Target
General Information of Target
| Target ID | LDTP00913 | |||||
|---|---|---|---|---|---|---|
| Target Name | Mitochondrial import inner membrane translocase subunit Tim8 A (TIMM8A) | |||||
| Gene Name | TIMM8A | |||||
| Gene ID | 1678 | |||||
| Synonyms |
DDP; DDP1; TIM8A; Mitochondrial import inner membrane translocase subunit Tim8 A; Deafness dystonia protein 1; X-linked deafness dystonia protein |
|||||
| 3D Structure | ||||||
| Sequence |
MDSSSSSSAAGLGAVDPQLQHFIEVETQKQRFQQLVHQMTELCWEKCMDKPGPKLDSRAE
ACFVNCVERFIDTSQFILNRLEQTQKSKPVFSESLSD |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Small Tim family
|
|||||
| Subcellular location |
Mitochondrion inner membrane
|
|||||
| Function |
Mitochondrial intermembrane chaperone that participates in the import and insertion of some multi-pass transmembrane proteins into the mitochondrial inner membrane. Also required for the transfer of beta-barrel precursors from the TOM complex to the sorting and assembly machinery (SAM complex) of the outer membrane. Acts as a chaperone-like protein that protects the hydrophobic precursors from aggregation and guide them through the mitochondrial intermembrane space. The TIMM8-TIMM13 complex mediates the import of proteins such as TIMM23, SLC25A12/ARALAR1 and SLC25A13/ARALAR2, while the predominant TIMM9-TIMM10 70 kDa complex mediates the import of much more proteins. Probably necessary for normal neurologic development.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
ONAyne Probe Info |
 |
K86(0.51) | LDD0274 | [1] | |
|
Acrolein Probe Info |
 |
C43(0.00); C66(0.00) | LDD0222 | [2] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [3] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [4] | |
|
TFBX Probe Info |
 |
C62(0.00); C66(0.00) | LDD0027 | [4] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [5] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [2] | |
|
NAIA_5 Probe Info |
 |
C66(0.00); C62(0.00); C47(0.00); C43(0.00) | LDD2223 | [6] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C106 Probe Info |
 |
24.76 | LDD1793 | [7] | |
|
C108 Probe Info |
 |
8.46 | LDD1795 | [7] | |
|
C232 Probe Info |
 |
37.27 | LDD1905 | [7] | |
|
C293 Probe Info |
 |
20.11 | LDD1963 | [7] | |
|
C296 Probe Info |
 |
14.32 | LDD1966 | [7] | |
|
C313 Probe Info |
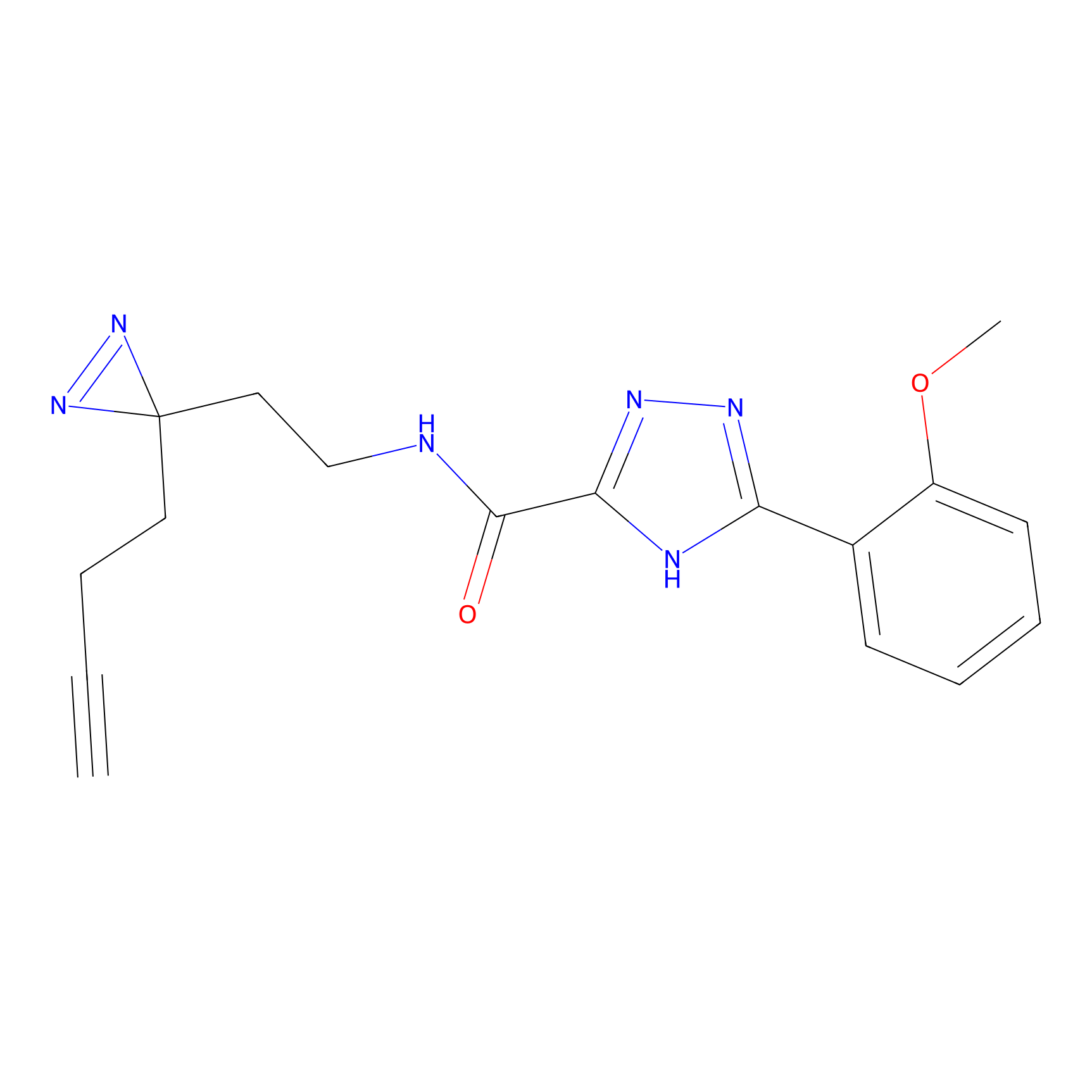 |
15.14 | LDD1980 | [7] | |
|
C314 Probe Info |
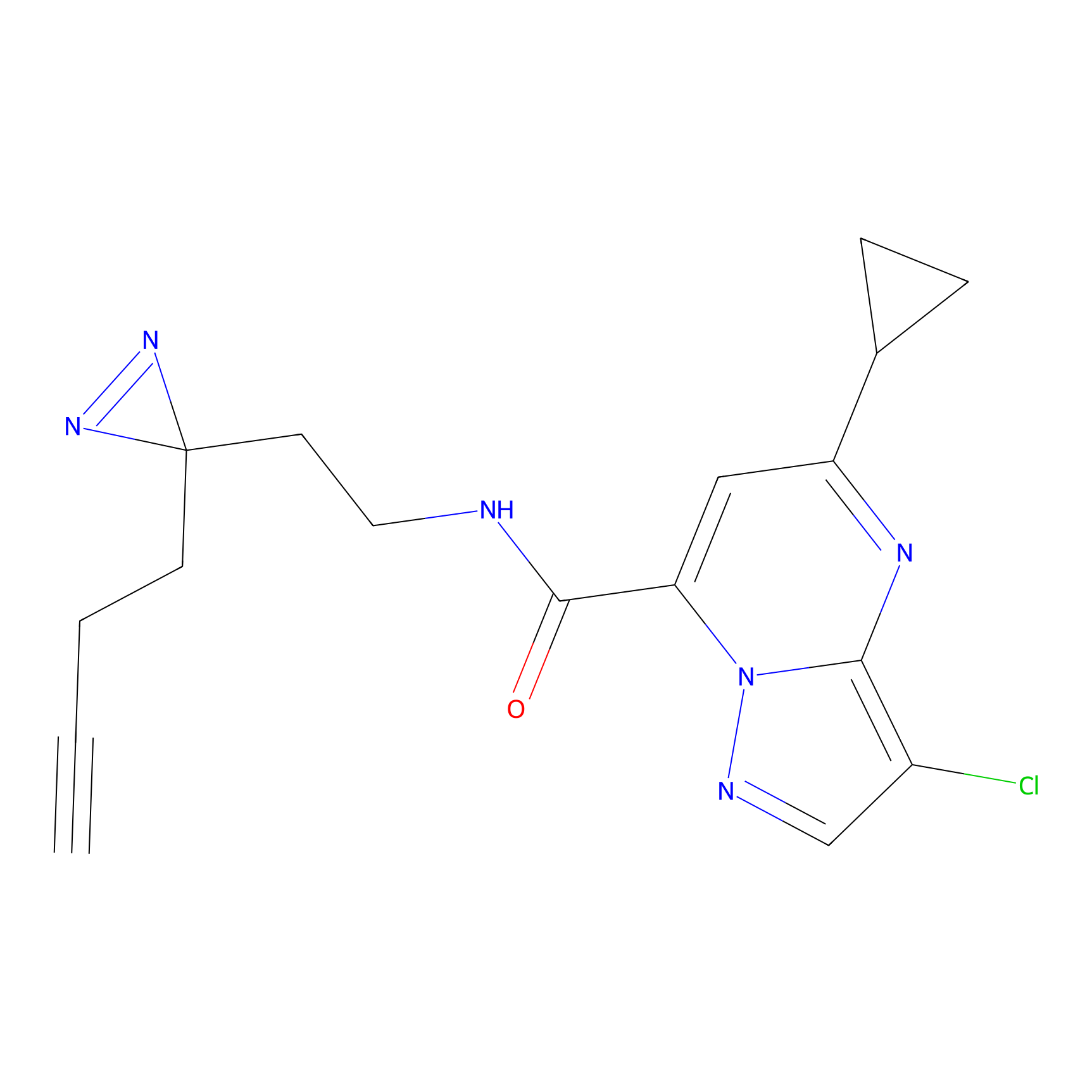 |
13.55 | LDD1981 | [7] | |
|
C343 Probe Info |
 |
12.04 | LDD2005 | [7] | |
|
C348 Probe Info |
 |
11.47 | LDD2009 | [7] | |
|
C349 Probe Info |
 |
10.70 | LDD2010 | [7] | |
|
C350 Probe Info |
 |
37.53 | LDD2011 | [7] | |
|
C354 Probe Info |
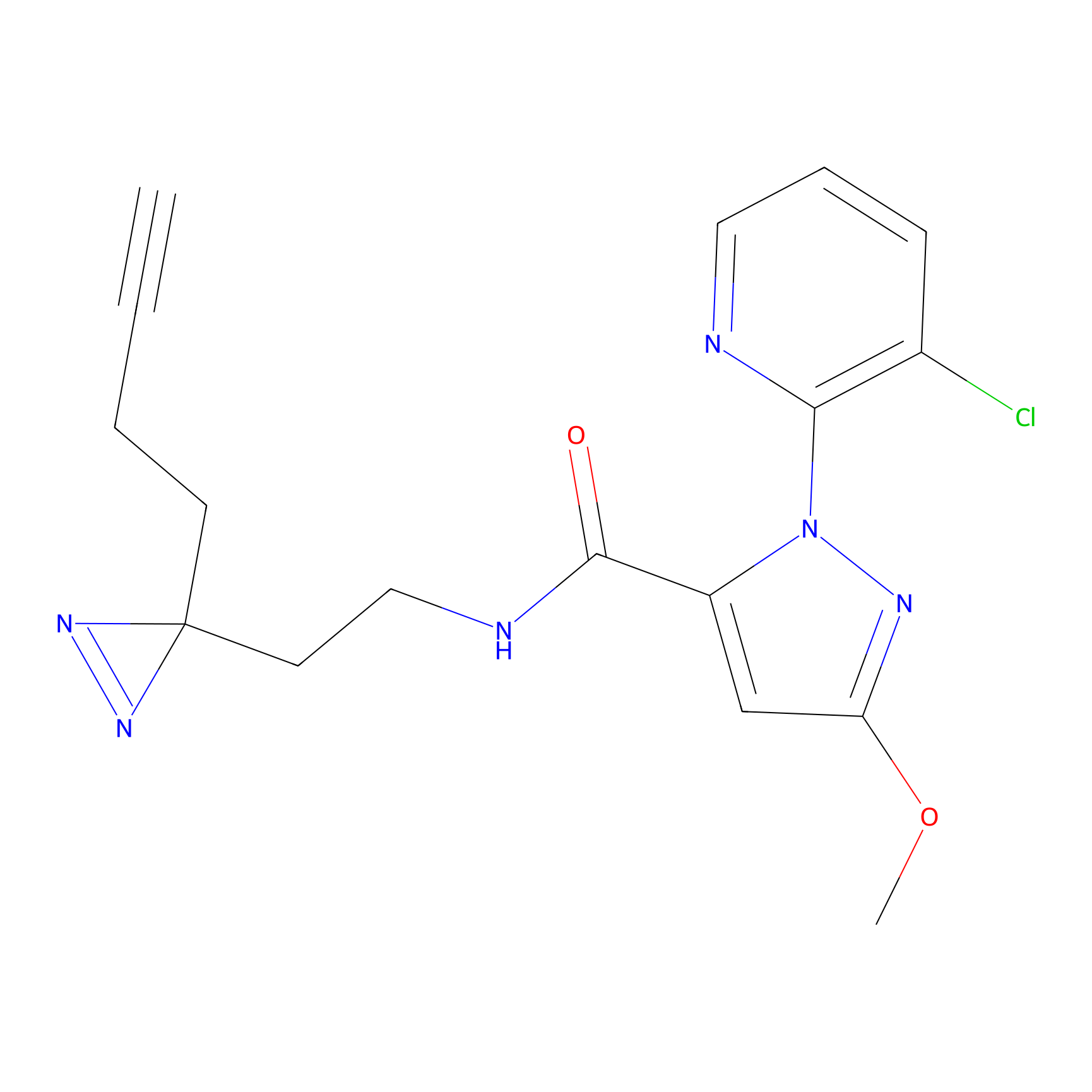 |
7.26 | LDD2015 | [7] | |
|
C362 Probe Info |
 |
25.28 | LDD2023 | [7] | |
Competitor(s) Related to This Target
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| mRNA export factor GLE1 (GLE1) | GLE1 family | Q53GS7 | |||
| Huntingtin (HTT) | Huntingtin family | P42858 | |||
| Wolframin (WFS1) | . | O76024 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| AT-rich interactive domain-containing protein 3A (ARID3A) | . | Q99856 | |||
Other
References
