Details of the Target
General Information of Target
| Target ID | LDTP14157 | |||||
|---|---|---|---|---|---|---|
| Target Name | RNA-binding protein 8A (RBM8A) | |||||
| Gene Name | RBM8A | |||||
| Gene ID | 9939 | |||||
| Synonyms |
RBM8; RNA-binding protein 8A; Binder of OVCA1-1; BOV-1; RNA-binding motif protein 8A; RNA-binding protein Y14; Ribonucleoprotein RBM8A |
|||||
| 3D Structure | ||||||
| Sequence |
MGNWVVNHWFSVLFLVVWLGLNVFLFVDAFLKYEKADKYYYTRKILGSTLACARASALCL
NFNSTLILLPVCRNLLSFLRGTCSFCSRTLRKQLDHNLTFHKLVAYMICLHTAIHIIAHL FNFDCYSRSRQATDGSLASILSSLSHDEKKGGSWLNPIQSRNTTVEYVTFTSIAGLTGVI MTIALILMVTSATEFIRRSYFEVFWYTHHLFIFYILGLGIHGIGGIVRGQTEESMNESHP RKCAESFEMWDDRDSHCRRPKFEGHPPESWKWILAPVILYICERILRFYRSQQKVVITKV VMHPSKVLELQMNKRGFSMEVGQYIFVNCPSISLLEWHPFTLTSAPEEDFFSIHIRAAGD WTENLIRAFEQQYSPIPRIEVDGPFGTASEDVFQYEVAVLVGAGIGVTPFASILKSIWYK FQCADHNLKTKKIYFYWICRETGAFSWFNNLLTSLEQEMEELGKVGFLNYRLFLTGWDSN IVGHAALNFDKATDIVTGLKQKTSFGRPMWDNEFSTIATSHPKSVVGVFLCGPRTLAKSL RKCCHRYSSLDPRKVQFYFNKENF |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
RBM8A family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Required for pre-mRNA splicing as component of the spliceosome. Core component of the splicing-dependent multiprotein exon junction complex (EJC) deposited at splice junctions on mRNAs. The EJC is a dynamic structure consisting of core proteins and several peripheral nuclear and cytoplasmic associated factors that join the complex only transiently either during EJC assembly or during subsequent mRNA metabolism. The EJC marks the position of the exon-exon junction in the mature mRNA for the gene expression machinery and the core components remain bound to spliced mRNAs throughout all stages of mRNA metabolism thereby influencing downstream processes including nuclear mRNA export, subcellular mRNA localization, translation efficiency and nonsense-mediated mRNA decay (NMD). The MAGOH-RBM8A heterodimer inhibits the ATPase activity of EIF4A3, thereby trapping the ATP-bound EJC core onto spliced mRNA in a stable conformation. The MAGOH-RBM8A heterodimer interacts with the EJC key regulator PYM1 leading to EJC disassembly in the cytoplasm and translation enhancement of EJC-bearing spliced mRNAs by recruiting them to the ribosomal 48S preinitiation complex. Its removal from cytoplasmic mRNAs requires translation initiation from EJC-bearing spliced mRNAs. Associates preferentially with mRNAs produced by splicing. Does not interact with pre-mRNAs, introns, or mRNAs produced from intronless cDNAs. Associates with both nuclear mRNAs and newly exported cytoplasmic mRNAs. The MAGOH-RBM8A heterodimer is a component of the nonsense mediated decay (NMD) pathway. Involved in the splicing modulation of BCL2L1/Bcl-X (and probably other apoptotic genes); specifically inhibits formation of proapoptotic isoforms such as Bcl-X(S); the function is different from the established EJC assembly.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
SAA-alkyne Probe Info |
 |
1.10 | LDD0252 | [1] | |
|
TH211 Probe Info |
 |
Y54(17.59) | LDD0260 | [2] | |
|
Probe 1 Probe Info |
 |
Y54(7,927.84); Y96(15.16); Y116(115.41) | LDD3495 | [3] | |
|
HHS-475 Probe Info |
 |
Y116(0.27); Y54(1.00) | LDD0264 | [4] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0221 | [5] | |
|
DBIA Probe Info |
 |
C149(0.96) | LDD0078 | [6] | |
|
m-APA Probe Info |
 |
N.A. | LDD2231 | [7] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [8] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [9] | |
|
IPIAA_H Probe Info |
 |
N.A. | LDD0030 | [10] | |
|
IPIAA_L Probe Info |
 |
N.A. | LDD0031 | [10] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [8] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [11] | |
|
HHS-465 Probe Info |
 |
Y54(0.00); Y116(0.00) | LDD2240 | [12] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
PARPYnD Probe Info |
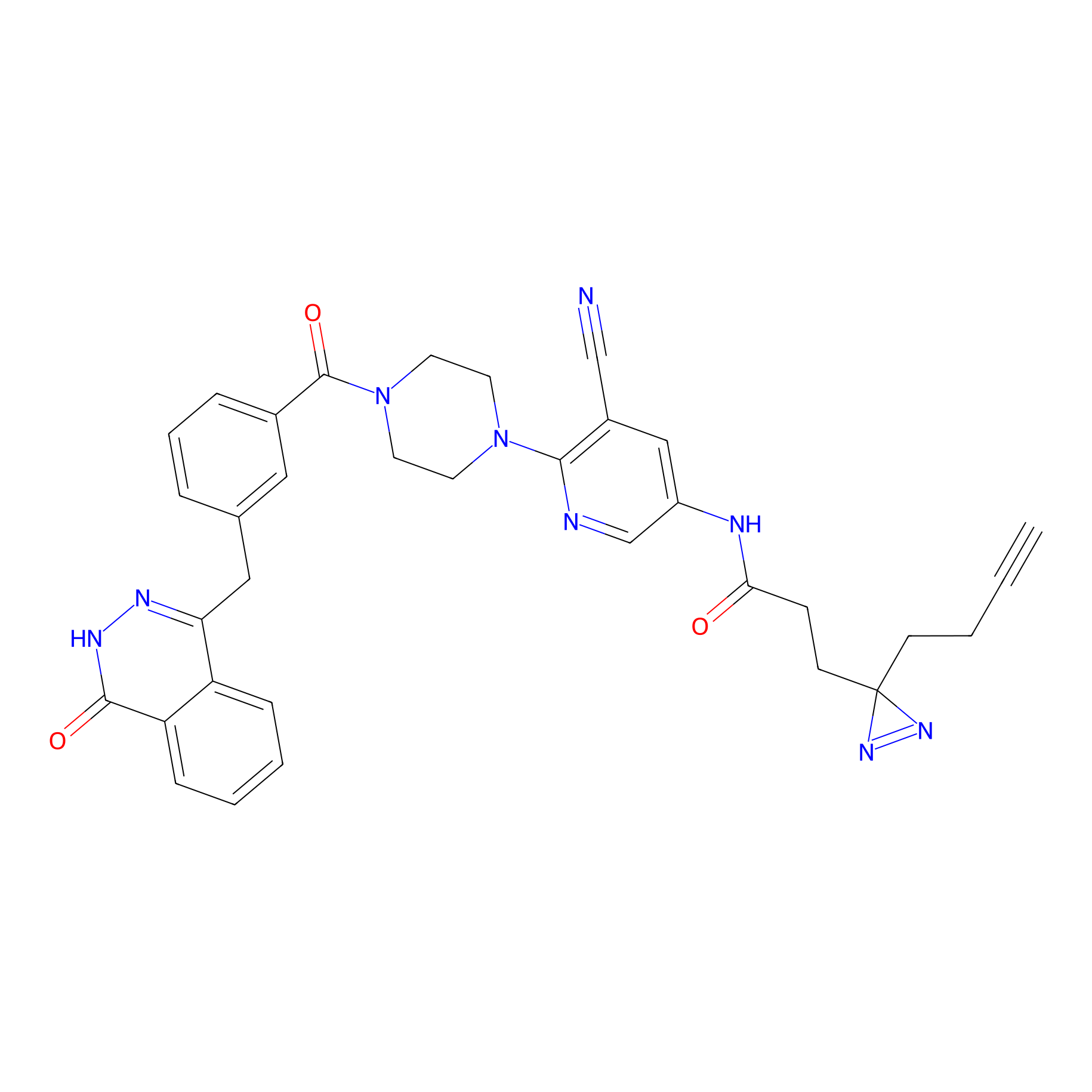 |
1.63 | LDD0376 | [13] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HCT 116 | C149(1.26) | LDD0531 | [6] |
| LDCM0215 | AC10 | HCT 116 | C149(1.00) | LDD0532 | [6] |
| LDCM0216 | AC100 | HCT 116 | C149(0.91) | LDD0533 | [6] |
| LDCM0217 | AC101 | HCT 116 | C149(0.89) | LDD0534 | [6] |
| LDCM0218 | AC102 | HCT 116 | C149(0.86) | LDD0535 | [6] |
| LDCM0219 | AC103 | HCT 116 | C149(0.78) | LDD0536 | [6] |
| LDCM0220 | AC104 | HCT 116 | C149(0.90) | LDD0537 | [6] |
| LDCM0221 | AC105 | HCT 116 | C149(0.84) | LDD0538 | [6] |
| LDCM0222 | AC106 | HCT 116 | C149(0.78) | LDD0539 | [6] |
| LDCM0223 | AC107 | HCT 116 | C149(0.73) | LDD0540 | [6] |
| LDCM0224 | AC108 | HCT 116 | C149(0.90) | LDD0541 | [6] |
| LDCM0225 | AC109 | HCT 116 | C149(0.76) | LDD0542 | [6] |
| LDCM0226 | AC11 | HCT 116 | C149(0.89) | LDD0543 | [6] |
| LDCM0227 | AC110 | HCT 116 | C149(0.90) | LDD0544 | [6] |
| LDCM0228 | AC111 | HCT 116 | C149(0.87) | LDD0545 | [6] |
| LDCM0229 | AC112 | HCT 116 | C149(0.84) | LDD0546 | [6] |
| LDCM0230 | AC113 | HCT 116 | C149(0.96) | LDD0547 | [6] |
| LDCM0231 | AC114 | HCT 116 | C149(0.99) | LDD0548 | [6] |
| LDCM0232 | AC115 | HCT 116 | C149(0.92) | LDD0549 | [6] |
| LDCM0233 | AC116 | HCT 116 | C149(0.84) | LDD0550 | [6] |
| LDCM0234 | AC117 | HCT 116 | C149(0.89) | LDD0551 | [6] |
| LDCM0235 | AC118 | HCT 116 | C149(0.87) | LDD0552 | [6] |
| LDCM0236 | AC119 | HCT 116 | C149(0.85) | LDD0553 | [6] |
| LDCM0237 | AC12 | HCT 116 | C149(1.01) | LDD0554 | [6] |
| LDCM0238 | AC120 | HCT 116 | C149(0.88) | LDD0555 | [6] |
| LDCM0239 | AC121 | HCT 116 | C149(0.95) | LDD0556 | [6] |
| LDCM0240 | AC122 | HCT 116 | C149(0.83) | LDD0557 | [6] |
| LDCM0241 | AC123 | HCT 116 | C149(0.96) | LDD0558 | [6] |
| LDCM0242 | AC124 | HCT 116 | C149(0.90) | LDD0559 | [6] |
| LDCM0243 | AC125 | HCT 116 | C149(0.86) | LDD0560 | [6] |
| LDCM0244 | AC126 | HCT 116 | C149(0.92) | LDD0561 | [6] |
| LDCM0245 | AC127 | HCT 116 | C149(0.84) | LDD0562 | [6] |
| LDCM0246 | AC128 | HCT 116 | C149(0.71) | LDD0563 | [6] |
| LDCM0247 | AC129 | HCT 116 | C149(1.51) | LDD0564 | [6] |
| LDCM0249 | AC130 | HCT 116 | C149(0.97) | LDD0566 | [6] |
| LDCM0250 | AC131 | HCT 116 | C149(1.67) | LDD0567 | [6] |
| LDCM0251 | AC132 | HCT 116 | C149(1.31) | LDD0568 | [6] |
| LDCM0252 | AC133 | HCT 116 | C149(1.11) | LDD0569 | [6] |
| LDCM0253 | AC134 | HCT 116 | C149(1.07) | LDD0570 | [6] |
| LDCM0254 | AC135 | HCT 116 | C149(0.93) | LDD0571 | [6] |
| LDCM0255 | AC136 | HCT 116 | C149(0.96) | LDD0572 | [6] |
| LDCM0256 | AC137 | HCT 116 | C149(1.19) | LDD0573 | [6] |
| LDCM0257 | AC138 | HCT 116 | C149(1.05) | LDD0574 | [6] |
| LDCM0258 | AC139 | HCT 116 | C149(1.13) | LDD0575 | [6] |
| LDCM0259 | AC14 | HCT 116 | C149(1.11) | LDD0576 | [6] |
| LDCM0260 | AC140 | HCT 116 | C149(0.97) | LDD0577 | [6] |
| LDCM0261 | AC141 | HCT 116 | C149(1.09) | LDD0578 | [6] |
| LDCM0262 | AC142 | HCT 116 | C149(1.36) | LDD0579 | [6] |
| LDCM0263 | AC143 | HCT 116 | C149(1.23) | LDD0580 | [6] |
| LDCM0264 | AC144 | HCT 116 | C149(0.90) | LDD0581 | [6] |
| LDCM0265 | AC145 | HCT 116 | C149(0.89) | LDD0582 | [6] |
| LDCM0266 | AC146 | HCT 116 | C149(0.97) | LDD0583 | [6] |
| LDCM0267 | AC147 | HCT 116 | C149(0.92) | LDD0584 | [6] |
| LDCM0268 | AC148 | HCT 116 | C149(0.51) | LDD0585 | [6] |
| LDCM0269 | AC149 | HCT 116 | C149(0.83) | LDD0586 | [6] |
| LDCM0270 | AC15 | HCT 116 | C149(1.05) | LDD0587 | [6] |
| LDCM0271 | AC150 | HCT 116 | C149(0.86) | LDD0588 | [6] |
| LDCM0272 | AC151 | HCT 116 | C149(1.00) | LDD0589 | [6] |
| LDCM0273 | AC152 | HCT 116 | C149(0.89) | LDD0590 | [6] |
| LDCM0274 | AC153 | HCT 116 | C149(0.44) | LDD0591 | [6] |
| LDCM0621 | AC154 | HCT 116 | C149(0.83) | LDD2158 | [6] |
| LDCM0622 | AC155 | HCT 116 | C149(1.02) | LDD2159 | [6] |
| LDCM0623 | AC156 | HCT 116 | C149(1.03) | LDD2160 | [6] |
| LDCM0624 | AC157 | HCT 116 | C149(1.11) | LDD2161 | [6] |
| LDCM0276 | AC17 | HCT 116 | C149(1.17) | LDD0593 | [6] |
| LDCM0277 | AC18 | HCT 116 | C149(0.95) | LDD0594 | [6] |
| LDCM0278 | AC19 | HCT 116 | C149(1.34) | LDD0595 | [6] |
| LDCM0279 | AC2 | HCT 116 | C149(1.15) | LDD0596 | [6] |
| LDCM0280 | AC20 | HCT 116 | C149(1.16) | LDD0597 | [6] |
| LDCM0281 | AC21 | HCT 116 | C149(1.18) | LDD0598 | [6] |
| LDCM0282 | AC22 | HCT 116 | C149(1.15) | LDD0599 | [6] |
| LDCM0283 | AC23 | HCT 116 | C149(1.06) | LDD0600 | [6] |
| LDCM0284 | AC24 | HCT 116 | C149(1.28) | LDD0601 | [6] |
| LDCM0285 | AC25 | HCT 116 | C149(1.17) | LDD0602 | [6] |
| LDCM0286 | AC26 | HCT 116 | C149(1.12) | LDD0603 | [6] |
| LDCM0287 | AC27 | HCT 116 | C149(1.33) | LDD0604 | [6] |
| LDCM0288 | AC28 | HCT 116 | C149(1.16) | LDD0605 | [6] |
| LDCM0289 | AC29 | HCT 116 | C149(1.11) | LDD0606 | [6] |
| LDCM0290 | AC3 | HCT 116 | C149(1.11) | LDD0607 | [6] |
| LDCM0291 | AC30 | HCT 116 | C149(1.29) | LDD0608 | [6] |
| LDCM0292 | AC31 | HCT 116 | C149(1.08) | LDD0609 | [6] |
| LDCM0293 | AC32 | HCT 116 | C149(1.31) | LDD0610 | [6] |
| LDCM0294 | AC33 | HCT 116 | C149(1.07) | LDD0611 | [6] |
| LDCM0295 | AC34 | HCT 116 | C149(1.13) | LDD0612 | [6] |
| LDCM0296 | AC35 | HCT 116 | C149(0.87) | LDD0613 | [6] |
| LDCM0297 | AC36 | HCT 116 | C149(0.89) | LDD0614 | [6] |
| LDCM0298 | AC37 | HCT 116 | C149(0.86) | LDD0615 | [6] |
| LDCM0299 | AC38 | HCT 116 | C149(0.90) | LDD0616 | [6] |
| LDCM0300 | AC39 | HCT 116 | C149(0.86) | LDD0617 | [6] |
| LDCM0301 | AC4 | HCT 116 | C149(1.15) | LDD0618 | [6] |
| LDCM0302 | AC40 | HCT 116 | C149(0.72) | LDD0619 | [6] |
| LDCM0303 | AC41 | HCT 116 | C149(0.77) | LDD0620 | [6] |
| LDCM0304 | AC42 | HCT 116 | C149(0.88) | LDD0621 | [6] |
| LDCM0305 | AC43 | HCT 116 | C149(0.93) | LDD0622 | [6] |
| LDCM0306 | AC44 | HCT 116 | C149(0.86) | LDD0623 | [6] |
| LDCM0307 | AC45 | HCT 116 | C149(0.76) | LDD0624 | [6] |
| LDCM0308 | AC46 | HCT 116 | C149(0.88) | LDD0625 | [6] |
| LDCM0309 | AC47 | HCT 116 | C149(0.92) | LDD0626 | [6] |
| LDCM0310 | AC48 | HCT 116 | C149(0.84) | LDD0627 | [6] |
| LDCM0311 | AC49 | HCT 116 | C149(1.02) | LDD0628 | [6] |
| LDCM0312 | AC5 | HCT 116 | C149(1.09) | LDD0629 | [6] |
| LDCM0313 | AC50 | HCT 116 | C149(0.88) | LDD0630 | [6] |
| LDCM0314 | AC51 | HCT 116 | C149(0.98) | LDD0631 | [6] |
| LDCM0315 | AC52 | HCT 116 | C149(0.92) | LDD0632 | [6] |
| LDCM0316 | AC53 | HCT 116 | C149(0.99) | LDD0633 | [6] |
| LDCM0317 | AC54 | HCT 116 | C149(1.06) | LDD0634 | [6] |
| LDCM0318 | AC55 | HCT 116 | C149(1.06) | LDD0635 | [6] |
| LDCM0319 | AC56 | HCT 116 | C149(1.07) | LDD0636 | [6] |
| LDCM0320 | AC57 | HCT 116 | C149(0.95) | LDD0637 | [6] |
| LDCM0321 | AC58 | HCT 116 | C149(1.00) | LDD0638 | [6] |
| LDCM0322 | AC59 | HCT 116 | C149(0.93) | LDD0639 | [6] |
| LDCM0323 | AC6 | HCT 116 | C149(1.01) | LDD0640 | [6] |
| LDCM0324 | AC60 | HCT 116 | C149(1.02) | LDD0641 | [6] |
| LDCM0325 | AC61 | HCT 116 | C149(0.96) | LDD0642 | [6] |
| LDCM0326 | AC62 | HCT 116 | C149(0.89) | LDD0643 | [6] |
| LDCM0327 | AC63 | HCT 116 | C149(0.96) | LDD0644 | [6] |
| LDCM0328 | AC64 | HCT 116 | C149(0.96) | LDD0645 | [6] |
| LDCM0329 | AC65 | HCT 116 | C149(1.04) | LDD0646 | [6] |
| LDCM0330 | AC66 | HCT 116 | C149(1.17) | LDD0647 | [6] |
| LDCM0331 | AC67 | HCT 116 | C149(0.70) | LDD0648 | [6] |
| LDCM0332 | AC68 | HCT 116 | C149(0.98) | LDD0649 | [6] |
| LDCM0333 | AC69 | HCT 116 | C149(0.85) | LDD0650 | [6] |
| LDCM0334 | AC7 | HCT 116 | C149(0.93) | LDD0651 | [6] |
| LDCM0335 | AC70 | HCT 116 | C149(0.65) | LDD0652 | [6] |
| LDCM0336 | AC71 | HCT 116 | C149(0.99) | LDD0653 | [6] |
| LDCM0337 | AC72 | HCT 116 | C149(0.97) | LDD0654 | [6] |
| LDCM0338 | AC73 | HCT 116 | C149(0.59) | LDD0655 | [6] |
| LDCM0339 | AC74 | HCT 116 | C149(0.67) | LDD0656 | [6] |
| LDCM0340 | AC75 | HCT 116 | C149(0.46) | LDD0657 | [6] |
| LDCM0341 | AC76 | HCT 116 | C149(0.79) | LDD0658 | [6] |
| LDCM0342 | AC77 | HCT 116 | C149(0.74) | LDD0659 | [6] |
| LDCM0343 | AC78 | HCT 116 | C149(1.00) | LDD0660 | [6] |
| LDCM0344 | AC79 | HCT 116 | C149(0.91) | LDD0661 | [6] |
| LDCM0345 | AC8 | HCT 116 | C149(0.95) | LDD0662 | [6] |
| LDCM0346 | AC80 | HCT 116 | C149(0.88) | LDD0663 | [6] |
| LDCM0347 | AC81 | HCT 116 | C149(0.88) | LDD0664 | [6] |
| LDCM0348 | AC82 | HCT 116 | C149(0.44) | LDD0665 | [6] |
| LDCM0349 | AC83 | HCT 116 | C149(0.78) | LDD0666 | [6] |
| LDCM0350 | AC84 | HCT 116 | C149(0.80) | LDD0667 | [6] |
| LDCM0351 | AC85 | HCT 116 | C149(0.62) | LDD0668 | [6] |
| LDCM0352 | AC86 | HCT 116 | C149(0.82) | LDD0669 | [6] |
| LDCM0353 | AC87 | HCT 116 | C149(0.87) | LDD0670 | [6] |
| LDCM0354 | AC88 | HCT 116 | C149(0.91) | LDD0671 | [6] |
| LDCM0355 | AC89 | HCT 116 | C149(0.83) | LDD0672 | [6] |
| LDCM0357 | AC90 | HCT 116 | C149(1.16) | LDD0674 | [6] |
| LDCM0358 | AC91 | HCT 116 | C149(0.61) | LDD0675 | [6] |
| LDCM0359 | AC92 | HCT 116 | C149(0.73) | LDD0676 | [6] |
| LDCM0360 | AC93 | HCT 116 | C149(0.85) | LDD0677 | [6] |
| LDCM0361 | AC94 | HCT 116 | C149(1.12) | LDD0678 | [6] |
| LDCM0362 | AC95 | HCT 116 | C149(0.99) | LDD0679 | [6] |
| LDCM0363 | AC96 | HCT 116 | C149(0.89) | LDD0680 | [6] |
| LDCM0364 | AC97 | HCT 116 | C149(0.77) | LDD0681 | [6] |
| LDCM0365 | AC98 | HCT 116 | C149(0.48) | LDD0682 | [6] |
| LDCM0366 | AC99 | HCT 116 | C149(0.80) | LDD0683 | [6] |
| LDCM0248 | AKOS034007472 | HCT 116 | C149(0.99) | LDD0565 | [6] |
| LDCM0356 | AKOS034007680 | HCT 116 | C149(1.06) | LDD0673 | [6] |
| LDCM0275 | AKOS034007705 | HCT 116 | C149(0.81) | LDD0592 | [6] |
| LDCM0020 | ARS-1620 | HCC44 | C149(0.96) | LDD0078 | [6] |
| LDCM0147 | AZ0108 | MDA-MB-468 | 1.63 | LDD0376 | [13] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [5] |
| LDCM0367 | CL1 | HCT 116 | C149(0.86) | LDD0684 | [6] |
| LDCM0368 | CL10 | HCT 116 | C149(0.97) | LDD0685 | [6] |
| LDCM0369 | CL100 | HCT 116 | C149(1.07) | LDD0686 | [6] |
| LDCM0370 | CL101 | HCT 116 | C149(0.99) | LDD0687 | [6] |
| LDCM0371 | CL102 | HCT 116 | C149(0.98) | LDD0688 | [6] |
| LDCM0372 | CL103 | HCT 116 | C149(1.04) | LDD0689 | [6] |
| LDCM0373 | CL104 | HCT 116 | C149(1.07) | LDD0690 | [6] |
| LDCM0374 | CL105 | HCT 116 | C149(0.98) | LDD0691 | [6] |
| LDCM0375 | CL106 | HCT 116 | C149(0.95) | LDD0692 | [6] |
| LDCM0376 | CL107 | HCT 116 | C149(1.13) | LDD0693 | [6] |
| LDCM0377 | CL108 | HCT 116 | C149(1.07) | LDD0694 | [6] |
| LDCM0378 | CL109 | HCT 116 | C149(0.97) | LDD0695 | [6] |
| LDCM0379 | CL11 | HCT 116 | C149(0.95) | LDD0696 | [6] |
| LDCM0380 | CL110 | HCT 116 | C149(1.03) | LDD0697 | [6] |
| LDCM0381 | CL111 | HCT 116 | C149(0.97) | LDD0698 | [6] |
| LDCM0382 | CL112 | HCT 116 | C149(1.15) | LDD0699 | [6] |
| LDCM0383 | CL113 | HCT 116 | C149(1.14) | LDD0700 | [6] |
| LDCM0384 | CL114 | HCT 116 | C149(1.09) | LDD0701 | [6] |
| LDCM0385 | CL115 | HCT 116 | C149(1.09) | LDD0702 | [6] |
| LDCM0386 | CL116 | HCT 116 | C149(1.02) | LDD0703 | [6] |
| LDCM0387 | CL117 | HCT 116 | C149(0.57) | LDD0704 | [6] |
| LDCM0388 | CL118 | HCT 116 | C149(0.81) | LDD0705 | [6] |
| LDCM0389 | CL119 | HCT 116 | C149(0.76) | LDD0706 | [6] |
| LDCM0390 | CL12 | HCT 116 | C149(1.06) | LDD0707 | [6] |
| LDCM0391 | CL120 | HCT 116 | C149(0.78) | LDD0708 | [6] |
| LDCM0392 | CL121 | HCT 116 | C149(0.97) | LDD0709 | [6] |
| LDCM0393 | CL122 | HCT 116 | C149(0.91) | LDD0710 | [6] |
| LDCM0394 | CL123 | HCT 116 | C149(0.87) | LDD0711 | [6] |
| LDCM0395 | CL124 | HCT 116 | C149(0.91) | LDD0712 | [6] |
| LDCM0396 | CL125 | HCT 116 | C149(0.97) | LDD0713 | [6] |
| LDCM0397 | CL126 | HCT 116 | C149(1.05) | LDD0714 | [6] |
| LDCM0398 | CL127 | HCT 116 | C149(1.04) | LDD0715 | [6] |
| LDCM0399 | CL128 | HCT 116 | C149(0.88) | LDD0716 | [6] |
| LDCM0400 | CL13 | HCT 116 | C149(1.02) | LDD0717 | [6] |
| LDCM0401 | CL14 | HCT 116 | C149(1.00) | LDD0718 | [6] |
| LDCM0402 | CL15 | HCT 116 | C149(0.89) | LDD0719 | [6] |
| LDCM0403 | CL16 | HCT 116 | C149(1.10) | LDD0720 | [6] |
| LDCM0404 | CL17 | HCT 116 | C149(0.90) | LDD0721 | [6] |
| LDCM0405 | CL18 | HCT 116 | C149(1.04) | LDD0722 | [6] |
| LDCM0406 | CL19 | HCT 116 | C149(1.07) | LDD0723 | [6] |
| LDCM0407 | CL2 | HCT 116 | C149(0.87) | LDD0724 | [6] |
| LDCM0408 | CL20 | HCT 116 | C149(1.15) | LDD0725 | [6] |
| LDCM0409 | CL21 | HCT 116 | C149(1.02) | LDD0726 | [6] |
| LDCM0410 | CL22 | HCT 116 | C149(0.71) | LDD0727 | [6] |
| LDCM0411 | CL23 | HCT 116 | C149(1.03) | LDD0728 | [6] |
| LDCM0412 | CL24 | HCT 116 | C149(1.04) | LDD0729 | [6] |
| LDCM0413 | CL25 | HCT 116 | C149(0.93) | LDD0730 | [6] |
| LDCM0414 | CL26 | HCT 116 | C149(1.05) | LDD0731 | [6] |
| LDCM0415 | CL27 | HCT 116 | C149(1.03) | LDD0732 | [6] |
| LDCM0416 | CL28 | HCT 116 | C149(0.99) | LDD0733 | [6] |
| LDCM0417 | CL29 | HCT 116 | C149(0.98) | LDD0734 | [6] |
| LDCM0418 | CL3 | HCT 116 | C149(0.99) | LDD0735 | [6] |
| LDCM0419 | CL30 | HCT 116 | C149(0.97) | LDD0736 | [6] |
| LDCM0420 | CL31 | HCT 116 | C149(1.15) | LDD0737 | [6] |
| LDCM0421 | CL32 | HCT 116 | C149(1.01) | LDD0738 | [6] |
| LDCM0422 | CL33 | HCT 116 | C149(0.91) | LDD0739 | [6] |
| LDCM0423 | CL34 | HCT 116 | C149(0.91) | LDD0740 | [6] |
| LDCM0424 | CL35 | HCT 116 | C149(1.10) | LDD0741 | [6] |
| LDCM0425 | CL36 | HCT 116 | C149(1.02) | LDD0742 | [6] |
| LDCM0426 | CL37 | HCT 116 | C149(1.02) | LDD0743 | [6] |
| LDCM0428 | CL39 | HCT 116 | C149(0.93) | LDD0745 | [6] |
| LDCM0429 | CL4 | HCT 116 | C149(0.91) | LDD0746 | [6] |
| LDCM0430 | CL40 | HCT 116 | C149(0.99) | LDD0747 | [6] |
| LDCM0431 | CL41 | HCT 116 | C149(1.05) | LDD0748 | [6] |
| LDCM0432 | CL42 | HCT 116 | C149(0.72) | LDD0749 | [6] |
| LDCM0433 | CL43 | HCT 116 | C149(1.05) | LDD0750 | [6] |
| LDCM0434 | CL44 | HCT 116 | C149(1.08) | LDD0751 | [6] |
| LDCM0435 | CL45 | HCT 116 | C149(1.03) | LDD0752 | [6] |
| LDCM0436 | CL46 | HCT 116 | C149(0.91) | LDD0753 | [6] |
| LDCM0437 | CL47 | HCT 116 | C149(0.99) | LDD0754 | [6] |
| LDCM0438 | CL48 | HCT 116 | C149(1.13) | LDD0755 | [6] |
| LDCM0439 | CL49 | HCT 116 | C149(1.19) | LDD0756 | [6] |
| LDCM0440 | CL5 | HCT 116 | C149(1.02) | LDD0757 | [6] |
| LDCM0441 | CL50 | HCT 116 | C149(0.95) | LDD0758 | [6] |
| LDCM0442 | CL51 | HCT 116 | C149(0.98) | LDD0759 | [6] |
| LDCM0443 | CL52 | HCT 116 | C149(0.96) | LDD0760 | [6] |
| LDCM0444 | CL53 | HCT 116 | C149(1.21) | LDD0761 | [6] |
| LDCM0445 | CL54 | HCT 116 | C149(0.94) | LDD0762 | [6] |
| LDCM0446 | CL55 | HCT 116 | C149(1.31) | LDD0763 | [6] |
| LDCM0447 | CL56 | HCT 116 | C149(1.00) | LDD0764 | [6] |
| LDCM0448 | CL57 | HCT 116 | C149(0.93) | LDD0765 | [6] |
| LDCM0449 | CL58 | HCT 116 | C149(1.06) | LDD0766 | [6] |
| LDCM0450 | CL59 | HCT 116 | C149(0.94) | LDD0767 | [6] |
| LDCM0451 | CL6 | HCT 116 | C149(0.96) | LDD0768 | [6] |
| LDCM0452 | CL60 | HCT 116 | C149(1.02) | LDD0769 | [6] |
| LDCM0453 | CL61 | HCT 116 | C149(1.03) | LDD0770 | [6] |
| LDCM0454 | CL62 | HCT 116 | C149(0.98) | LDD0771 | [6] |
| LDCM0455 | CL63 | HCT 116 | C149(0.90) | LDD0772 | [6] |
| LDCM0456 | CL64 | HCT 116 | C149(0.95) | LDD0773 | [6] |
| LDCM0457 | CL65 | HCT 116 | C149(1.04) | LDD0774 | [6] |
| LDCM0458 | CL66 | HCT 116 | C149(0.98) | LDD0775 | [6] |
| LDCM0459 | CL67 | HCT 116 | C149(0.97) | LDD0776 | [6] |
| LDCM0460 | CL68 | HCT 116 | C149(0.90) | LDD0777 | [6] |
| LDCM0461 | CL69 | HCT 116 | C149(1.01) | LDD0778 | [6] |
| LDCM0462 | CL7 | HCT 116 | C149(1.12) | LDD0779 | [6] |
| LDCM0463 | CL70 | HCT 116 | C149(0.97) | LDD0780 | [6] |
| LDCM0464 | CL71 | HCT 116 | C149(0.92) | LDD0781 | [6] |
| LDCM0465 | CL72 | HCT 116 | C149(0.90) | LDD0782 | [6] |
| LDCM0466 | CL73 | HCT 116 | C149(0.97) | LDD0783 | [6] |
| LDCM0467 | CL74 | HCT 116 | C149(0.90) | LDD0784 | [6] |
| LDCM0469 | CL76 | HCT 116 | C149(1.07) | LDD0786 | [6] |
| LDCM0470 | CL77 | HCT 116 | C149(0.96) | LDD0787 | [6] |
| LDCM0471 | CL78 | HCT 116 | C149(1.05) | LDD0788 | [6] |
| LDCM0472 | CL79 | HCT 116 | C149(1.13) | LDD0789 | [6] |
| LDCM0473 | CL8 | HCT 116 | C149(0.74) | LDD0790 | [6] |
| LDCM0474 | CL80 | HCT 116 | C149(1.13) | LDD0791 | [6] |
| LDCM0475 | CL81 | HCT 116 | C149(1.07) | LDD0792 | [6] |
| LDCM0476 | CL82 | HCT 116 | C149(0.92) | LDD0793 | [6] |
| LDCM0477 | CL83 | HCT 116 | C149(0.91) | LDD0794 | [6] |
| LDCM0478 | CL84 | HCT 116 | C149(0.73) | LDD0795 | [6] |
| LDCM0479 | CL85 | HCT 116 | C149(1.08) | LDD0796 | [6] |
| LDCM0480 | CL86 | HCT 116 | C149(1.12) | LDD0797 | [6] |
| LDCM0481 | CL87 | HCT 116 | C149(1.01) | LDD0798 | [6] |
| LDCM0482 | CL88 | HCT 116 | C149(0.98) | LDD0799 | [6] |
| LDCM0483 | CL89 | HCT 116 | C149(0.76) | LDD0800 | [6] |
| LDCM0484 | CL9 | HCT 116 | C149(0.98) | LDD0801 | [6] |
| LDCM0485 | CL90 | HCT 116 | C149(0.97) | LDD0802 | [6] |
| LDCM0486 | CL91 | HCT 116 | C149(0.91) | LDD0803 | [6] |
| LDCM0487 | CL92 | HCT 116 | C149(0.88) | LDD0804 | [6] |
| LDCM0488 | CL93 | HCT 116 | C149(0.96) | LDD0805 | [6] |
| LDCM0489 | CL94 | HCT 116 | C149(0.98) | LDD0806 | [6] |
| LDCM0490 | CL95 | HCT 116 | C149(0.88) | LDD0807 | [6] |
| LDCM0491 | CL96 | HCT 116 | C149(0.95) | LDD0808 | [6] |
| LDCM0492 | CL97 | HCT 116 | C149(1.13) | LDD0809 | [6] |
| LDCM0493 | CL98 | HCT 116 | C149(1.08) | LDD0810 | [6] |
| LDCM0494 | CL99 | HCT 116 | C149(1.12) | LDD0811 | [6] |
| LDCM0495 | E2913 | HEK-293T | C149(1.02) | LDD1698 | [14] |
| LDCM0468 | Fragment33 | HCT 116 | C149(0.93) | LDD0785 | [6] |
| LDCM0427 | Fragment51 | HCT 116 | C149(0.89) | LDD0744 | [6] |
| LDCM0116 | HHS-0101 | DM93 | Y116(0.27); Y54(1.00) | LDD0264 | [4] |
| LDCM0117 | HHS-0201 | DM93 | Y54(0.69); Y116(0.96) | LDD0265 | [4] |
| LDCM0118 | HHS-0301 | DM93 | Y116(0.38); Y54(0.77) | LDD0266 | [4] |
| LDCM0119 | HHS-0401 | DM93 | Y116(0.45); Y54(0.78) | LDD0267 | [4] |
| LDCM0120 | HHS-0701 | DM93 | Y116(0.50); Y54(1.59) | LDD0268 | [4] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [5] |
| LDCM0022 | KB02 | HCT 116 | C149(1.51) | LDD0080 | [6] |
| LDCM0023 | KB03 | HCT 116 | C149(0.90) | LDD0081 | [6] |
| LDCM0024 | KB05 | HCT 116 | C149(0.91) | LDD0082 | [6] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [5] |
| LDCM0131 | RA190 | MM1.R | C149(1.37) | LDD0304 | [15] |
| LDCM0021 | THZ1 | HCT 116 | C149(0.97) | LDD2173 | [6] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Protein mago nashi homolog (MAGOH) | Mago nashi family | P61326 | |||
Other
References
