Details of the Target
General Information of Target
| Target ID | LDTP12312 | |||||
|---|---|---|---|---|---|---|
| Target Name | Ras-related protein Rab-18 (RAB18) | |||||
| Gene Name | RAB18 | |||||
| Gene ID | 22931 | |||||
| Synonyms |
Ras-related protein Rab-18 |
|||||
| 3D Structure | ||||||
| Sequence |
MAGALAGLAAGLQVPRVAPSPDSDSDTDSEDPSLRRSAGGLLRSQVIHSGHFMVSSPHSD
SLPRRRDQEGSVGPSDFGPRSIDPTLTRLFECLSLAYSGKLVSPKWKNFKGLKLLCRDKI RLNNAIWRAWYIQYVKRRKSPVCGFVTPLQGPEADAHRKPEAVVLEGNYWKRRIEVVMRE YHKWRIYYKKRLRKPSREDDLLAPKQAEGRWPPPEQWCKQLFSSVVPVLLGDPEEEPGGR QLLDLNCFLSDISDTLFTMTQSGPSPLQLPPEDAYVGNADMIQPDLTPLQPSLDDFMDIS DFFTNSRLPQPPMPSNFPEPPSFSPVVDSLFSSGTLGPEVPPASSAMTHLSGHSRLQARN SCPGPLDSSAFLSSDFLLPEDPKPRLPPPPVPPPLLHYPPPAKVPGLEPCPPPPFPPMAP PTALLQEEPLFSPRFPFPTVPPAPGVSPLPAPAAFPPTPQSVPSPAPTPFPIELLPLGYS EPAFGPCFSMPRGKPPAPSPRGQKASPPTLAPATASPPTTAGSNNPCLTQLLTAAKPEQA LEPPLVSSTLLRSPGSPQETVPEFPCTFLPPTPAPTPPRPPPGPATLAPSRPLLVPKAER LSPPAPSGSERRLSGDLSSMPGPGTLSVRVSPPQPILSRGRPDSNKTENRRITHISAEQK RRFNIKLGFDTLHGLVSTLSAQPSLKVSKATTLQKTAEYILMLQQERAGLQEEAQQLRDE IEELNAAINLCQQQLPATGVPITHQRFDQMRDMFDDYVRTRTLHNWKFWVFSILIRPLFE SFNGMVSTASVHTLRQTSLAWLDQYCSLPALRPTVLNSLRQLGTSTSILTDPGRIPEQAT RAVTEGTLGKPL |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Small GTPase superfamily, Rab family
|
|||||
| Subcellular location |
Apical cell membrane
|
|||||
| Function |
The small GTPases Rab are key regulators of intracellular membrane trafficking, from the formation of transport vesicles to their fusion with membranes. Rabs cycle between an inactive GDP-bound form and an active GTP-bound form that is able to recruit to membranes different sets of downstream effectors directly responsible for vesicle formation, movement, tethering and fusion. Required for the localization of ZFYVE1 to lipid droplets and for its function in mediating the formation of endoplasmic reticulum-lipid droplets (ER-LD) contacts. Also required for maintaining endoplasmic reticulum structure. Plays a role in apical endocytosis/recycling. Plays a key role in eye and brain development and neurodegeneration.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| KARPAS422 | SNV: p.Q67P | . | |||
| REH | SNV: p.G200V | . | |||
| SW1783 | SNV: p.D125N | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
STPyne Probe Info |
 |
K138(10.00); K142(10.00); K48(7.69); K58(8.04) | LDD0277 | [2] | |
|
IPM Probe Info |
 |
C160(0.00); C155(0.00); C110(0.00) | LDD0241 | [3] | |
|
DBIA Probe Info |
 |
C139(1.56) | LDD3310 | [4] | |
|
DA-P3 Probe Info |
 |
5.31 | LDD0179 | [5] | |
|
HPAP Probe Info |
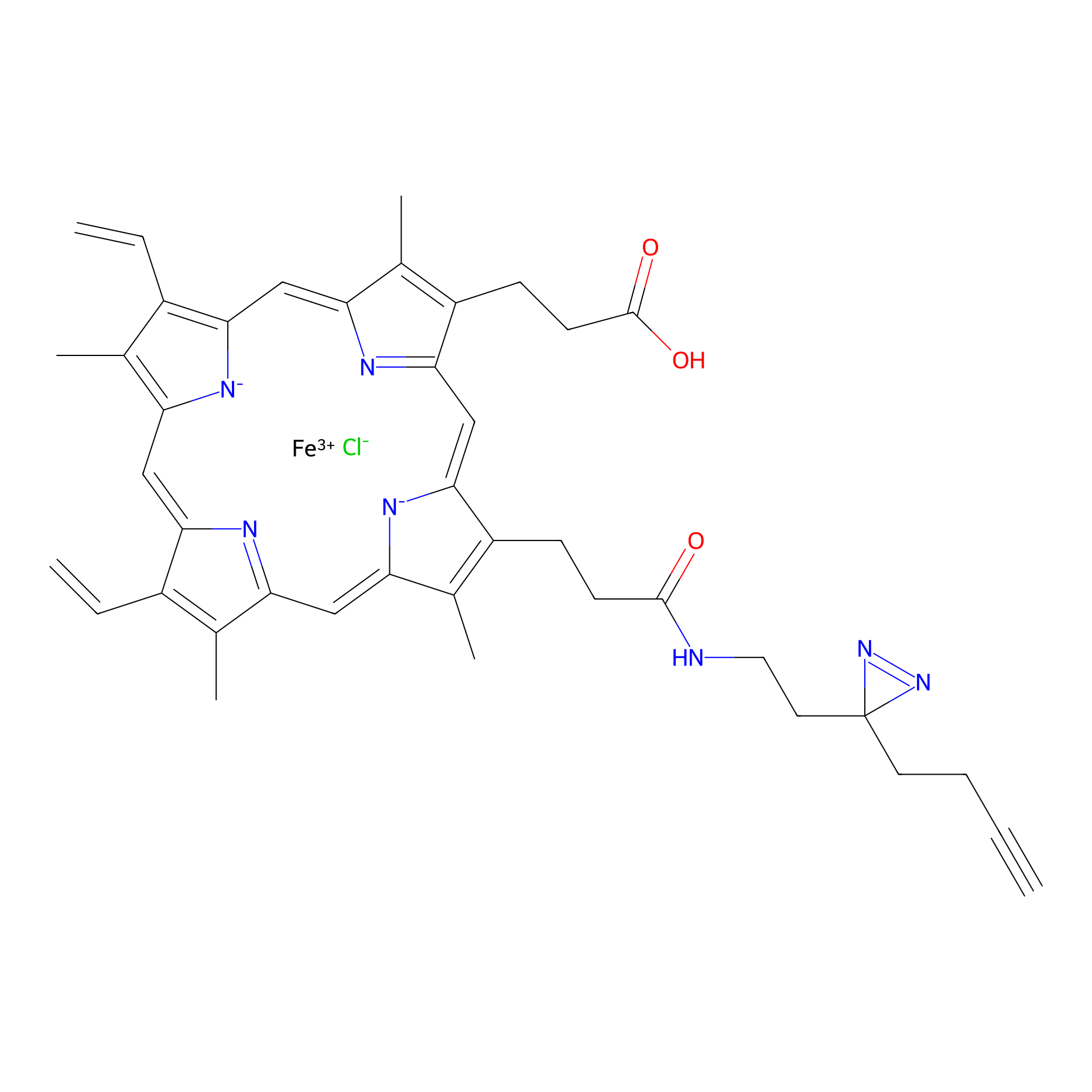 |
4.32 | LDD0064 | [6] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [7] | |
|
IA-alkyne Probe Info |
 |
C155(0.00); C110(0.00) | LDD0032 | [8] | |
|
IPIAA_H Probe Info |
 |
N.A. | LDD0030 | [9] | |
|
IPIAA_L Probe Info |
 |
C110(0.00); C155(0.00) | LDD0031 | [9] | |
|
Lodoacetamide azide Probe Info |
 |
C110(0.00); C155(0.00); C160(0.00) | LDD0037 | [7] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [10] | |
|
NAIA_4 Probe Info |
 |
C110(0.00); C155(0.00) | LDD2226 | [11] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [12] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [13] | |
|
Acrolein Probe Info |
 |
C155(0.00); C160(0.00) | LDD0217 | [14] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [14] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [3] | |
|
AOyne Probe Info |
 |
14.20 | LDD0443 | [15] | |
|
NAIA_5 Probe Info |
 |
C110(0.00); C160(0.00); C155(0.00) | LDD2223 | [11] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C235 Probe Info |
 |
20.25 | LDD1908 | [16] | |
|
C285 Probe Info |
 |
20.39 | LDD1955 | [16] | |
|
FFF probe11 Probe Info |
 |
9.81 | LDD0472 | [17] | |
|
FFF probe13 Probe Info |
 |
5.39 | LDD0476 | [17] | |
|
FFF probe3 Probe Info |
 |
20.00 | LDD0464 | [17] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HEK-293T | C110(1.04); C155(0.98); C160(0.96) | LDD1507 | [18] |
| LDCM0215 | AC10 | HEK-293T | C110(1.03); C155(0.91); C160(0.95) | LDD1508 | [18] |
| LDCM0226 | AC11 | HEK-293T | C110(0.85); C155(0.95); C160(1.08) | LDD1509 | [18] |
| LDCM0237 | AC12 | HEK-293T | C110(0.96); C155(1.09); C160(1.12) | LDD1510 | [18] |
| LDCM0259 | AC14 | HEK-293T | C110(0.95); C155(0.97); C160(0.93) | LDD1512 | [18] |
| LDCM0270 | AC15 | HEK-293T | C110(1.00); C160(1.06) | LDD1513 | [18] |
| LDCM0276 | AC17 | HEK-293T | C110(0.83); C155(1.05); C160(0.98) | LDD1515 | [18] |
| LDCM0277 | AC18 | HEK-293T | C110(1.00); C155(0.90); C160(0.92) | LDD1516 | [18] |
| LDCM0278 | AC19 | HEK-293T | C110(0.79); C155(0.80); C160(1.05) | LDD1517 | [18] |
| LDCM0279 | AC2 | HEK-293T | C110(1.02); C155(0.90); C160(1.01) | LDD1518 | [18] |
| LDCM0280 | AC20 | HEK-293T | C110(0.95); C155(1.06); C160(1.18) | LDD1519 | [18] |
| LDCM0281 | AC21 | HEK-293T | C110(1.04); C155(0.94); C160(1.03) | LDD1520 | [18] |
| LDCM0282 | AC22 | HEK-293T | C110(1.04); C155(1.01); C160(1.00) | LDD1521 | [18] |
| LDCM0283 | AC23 | HEK-293T | C110(1.07); C160(1.05) | LDD1522 | [18] |
| LDCM0284 | AC24 | HEK-293T | C110(1.02); C155(0.97); C160(1.18); C155(1.22) | LDD1523 | [18] |
| LDCM0285 | AC25 | HEK-293T | C110(0.97); C155(1.09); C160(0.99) | LDD1524 | [18] |
| LDCM0286 | AC26 | HEK-293T | C110(1.05); C155(0.98); C160(0.87) | LDD1525 | [18] |
| LDCM0287 | AC27 | HEK-293T | C110(0.82); C155(0.91); C160(0.97) | LDD1526 | [18] |
| LDCM0288 | AC28 | HEK-293T | C110(0.95); C155(1.11); C160(0.95) | LDD1527 | [18] |
| LDCM0289 | AC29 | HEK-293T | C110(1.12); C155(1.01); C160(0.98) | LDD1528 | [18] |
| LDCM0290 | AC3 | HEK-293T | C110(0.87); C155(1.02); C160(0.98) | LDD1529 | [18] |
| LDCM0291 | AC30 | HEK-293T | C110(0.99); C155(0.93); C160(1.02) | LDD1530 | [18] |
| LDCM0292 | AC31 | HEK-293T | C110(0.93); C160(0.98) | LDD1531 | [18] |
| LDCM0293 | AC32 | HEK-293T | C110(1.03); C155(1.00); C160(1.24); C155(1.02) | LDD1532 | [18] |
| LDCM0294 | AC33 | HEK-293T | C110(0.97); C155(1.01); C160(0.99) | LDD1533 | [18] |
| LDCM0295 | AC34 | HEK-293T | C110(1.07); C155(0.94); C160(0.93) | LDD1534 | [18] |
| LDCM0296 | AC35 | HEK-293T | C110(0.80); C155(0.99); C160(1.06) | LDD1535 | [18] |
| LDCM0297 | AC36 | HEK-293T | C110(0.97); C155(1.06); C160(1.00) | LDD1536 | [18] |
| LDCM0298 | AC37 | HEK-293T | C110(1.06); C155(1.00); C160(1.00) | LDD1537 | [18] |
| LDCM0299 | AC38 | HEK-293T | C110(1.00); C155(0.96); C160(0.94) | LDD1538 | [18] |
| LDCM0300 | AC39 | HEK-293T | C110(0.92); C160(0.97) | LDD1539 | [18] |
| LDCM0301 | AC4 | HEK-293T | C110(1.09); C155(0.91); C160(1.03) | LDD1540 | [18] |
| LDCM0302 | AC40 | HEK-293T | C110(1.00); C155(0.99); C160(1.06); C155(1.09) | LDD1541 | [18] |
| LDCM0303 | AC41 | HEK-293T | C110(1.08); C155(1.07); C160(1.02) | LDD1542 | [18] |
| LDCM0304 | AC42 | HEK-293T | C110(1.00); C155(0.91); C160(0.92) | LDD1543 | [18] |
| LDCM0305 | AC43 | HEK-293T | C110(0.87); C155(0.96); C160(0.95) | LDD1544 | [18] |
| LDCM0306 | AC44 | HEK-293T | C110(1.06); C155(1.01); C160(1.17) | LDD1545 | [18] |
| LDCM0307 | AC45 | HEK-293T | C110(1.00); C155(0.97); C160(1.01) | LDD1546 | [18] |
| LDCM0308 | AC46 | HEK-293T | C110(1.07); C155(1.00); C160(0.89) | LDD1547 | [18] |
| LDCM0309 | AC47 | HEK-293T | C110(1.03); C160(0.95) | LDD1548 | [18] |
| LDCM0310 | AC48 | HEK-293T | C110(0.92); C155(0.98); C160(1.16); C155(1.18) | LDD1549 | [18] |
| LDCM0311 | AC49 | HEK-293T | C110(1.10); C155(0.98); C160(0.96) | LDD1550 | [18] |
| LDCM0312 | AC5 | HEK-293T | C110(1.08); C155(0.85); C160(0.97) | LDD1551 | [18] |
| LDCM0313 | AC50 | HEK-293T | C110(1.13); C155(0.80); C160(0.91) | LDD1552 | [18] |
| LDCM0314 | AC51 | HEK-293T | C110(0.85); C155(0.89); C160(1.06) | LDD1553 | [18] |
| LDCM0315 | AC52 | HEK-293T | C110(1.12); C155(1.01); C160(1.03) | LDD1554 | [18] |
| LDCM0316 | AC53 | HEK-293T | C110(1.01); C155(0.96); C160(1.02) | LDD1555 | [18] |
| LDCM0317 | AC54 | HEK-293T | C110(1.06); C155(0.85); C160(0.91) | LDD1556 | [18] |
| LDCM0318 | AC55 | HEK-293T | C110(1.14); C160(1.01) | LDD1557 | [18] |
| LDCM0319 | AC56 | HEK-293T | C110(0.93); C155(1.00); C160(1.15); C155(1.33) | LDD1558 | [18] |
| LDCM0320 | AC57 | HEK-293T | C110(1.21); C155(1.19); C160(1.00) | LDD1559 | [18] |
| LDCM0321 | AC58 | HEK-293T | C110(1.01); C155(0.95); C160(0.91) | LDD1560 | [18] |
| LDCM0322 | AC59 | HEK-293T | C110(0.86); C155(1.06); C160(1.06) | LDD1561 | [18] |
| LDCM0323 | AC6 | HEK-293T | C110(1.19); C155(1.00); C160(0.92) | LDD1562 | [18] |
| LDCM0324 | AC60 | HEK-293T | C110(0.97); C155(0.96); C160(0.99) | LDD1563 | [18] |
| LDCM0325 | AC61 | HEK-293T | C110(1.04); C155(1.03); C160(1.01) | LDD1564 | [18] |
| LDCM0326 | AC62 | HEK-293T | C110(1.01); C155(1.00); C160(0.97) | LDD1565 | [18] |
| LDCM0327 | AC63 | HEK-293T | C110(0.99); C160(0.96) | LDD1566 | [18] |
| LDCM0328 | AC64 | HEK-293T | C110(0.99); C155(1.07); C160(1.11); C155(1.33) | LDD1567 | [18] |
| LDCM0334 | AC7 | HEK-293T | C110(1.10); C160(0.95) | LDD1568 | [18] |
| LDCM0345 | AC8 | HEK-293T | C110(0.99); C155(1.09); C160(1.11); C155(1.18) | LDD1569 | [18] |
| LDCM0248 | AKOS034007472 | HEK-293T | C110(1.02); C155(0.97); C160(1.01) | LDD1511 | [18] |
| LDCM0356 | AKOS034007680 | HEK-293T | C110(1.00); C155(1.02); C160(1.00) | LDD1570 | [18] |
| LDCM0275 | AKOS034007705 | HEK-293T | C110(1.09); C155(1.07); C160(1.09); C155(0.94) | LDD1514 | [18] |
| LDCM0156 | Aniline | NCI-H1299 | 12.60 | LDD0403 | [1] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [14] |
| LDCM0632 | CL-Sc | Hep-G2 | C155(0.98) | LDD2227 | [11] |
| LDCM0367 | CL1 | HEK-293T | C110(0.91); C155(1.02); C160(0.94) | LDD1571 | [18] |
| LDCM0368 | CL10 | HEK-293T | C110(1.28); C155(1.09); C160(0.97) | LDD1572 | [18] |
| LDCM0369 | CL100 | HEK-293T | C110(0.98); C155(0.97); C160(0.90) | LDD1573 | [18] |
| LDCM0370 | CL101 | HEK-293T | C110(0.93); C155(1.01); C160(0.99) | LDD1574 | [18] |
| LDCM0371 | CL102 | HEK-293T | C110(1.21); C155(0.99); C160(0.95) | LDD1575 | [18] |
| LDCM0372 | CL103 | HEK-293T | C110(0.96); C155(0.97); C160(1.09) | LDD1576 | [18] |
| LDCM0373 | CL104 | HEK-293T | C110(1.08); C155(1.06); C160(0.94) | LDD1577 | [18] |
| LDCM0374 | CL105 | HEK-293T | C110(0.86); C155(1.00); C160(1.02) | LDD1578 | [18] |
| LDCM0375 | CL106 | HEK-293T | C110(1.21); C155(1.07); C160(1.05) | LDD1579 | [18] |
| LDCM0376 | CL107 | HEK-293T | C110(0.89); C155(0.91); C160(1.01) | LDD1580 | [18] |
| LDCM0377 | CL108 | HEK-293T | C110(0.95); C155(0.93); C160(0.98) | LDD1581 | [18] |
| LDCM0378 | CL109 | HEK-293T | C110(0.93); C155(1.00); C160(1.00) | LDD1582 | [18] |
| LDCM0379 | CL11 | HEK-293T | C110(1.08); C160(1.08) | LDD1583 | [18] |
| LDCM0380 | CL110 | HEK-293T | C110(1.05); C155(1.02); C160(0.94) | LDD1584 | [18] |
| LDCM0381 | CL111 | HEK-293T | C110(0.86); C155(0.89); C160(0.91) | LDD1585 | [18] |
| LDCM0382 | CL112 | HEK-293T | C110(1.03); C155(1.01); C160(0.95) | LDD1586 | [18] |
| LDCM0383 | CL113 | HEK-293T | C110(0.97); C155(0.96); C160(0.93) | LDD1587 | [18] |
| LDCM0384 | CL114 | HEK-293T | C110(1.18); C155(0.59); C160(0.89) | LDD1588 | [18] |
| LDCM0385 | CL115 | HEK-293T | C110(0.81); C155(0.93); C160(0.96) | LDD1589 | [18] |
| LDCM0386 | CL116 | HEK-293T | C110(1.11); C155(1.05); C160(0.90) | LDD1590 | [18] |
| LDCM0387 | CL117 | HEK-293T | C110(1.03); C155(0.93); C160(1.02) | LDD1591 | [18] |
| LDCM0388 | CL118 | HEK-293T | C110(1.29); C155(0.97); C160(0.97) | LDD1592 | [18] |
| LDCM0389 | CL119 | HEK-293T | C110(0.82); C155(0.89); C160(0.99) | LDD1593 | [18] |
| LDCM0390 | CL12 | HEK-293T | C110(0.96); C155(1.07); C160(1.40); C155(1.10) | LDD1594 | [18] |
| LDCM0391 | CL120 | HEK-293T | C110(1.04); C155(0.94); C160(0.94) | LDD1595 | [18] |
| LDCM0392 | CL121 | HEK-293T | C110(0.93); C155(1.02); C160(0.96) | LDD1596 | [18] |
| LDCM0393 | CL122 | HEK-293T | C110(1.29); C155(0.87); C160(0.95) | LDD1597 | [18] |
| LDCM0394 | CL123 | HEK-293T | C110(0.96); C155(0.95); C160(0.90) | LDD1598 | [18] |
| LDCM0395 | CL124 | HEK-293T | C110(1.05); C155(0.81); C160(0.94) | LDD1599 | [18] |
| LDCM0396 | CL125 | HEK-293T | C110(0.95); C155(1.09); C160(0.96) | LDD1600 | [18] |
| LDCM0397 | CL126 | HEK-293T | C110(1.18); C155(1.07); C160(1.00) | LDD1601 | [18] |
| LDCM0398 | CL127 | HEK-293T | C110(0.83); C155(0.91); C160(0.93) | LDD1602 | [18] |
| LDCM0399 | CL128 | HEK-293T | C110(1.00); C155(1.00); C160(0.97) | LDD1603 | [18] |
| LDCM0400 | CL13 | HEK-293T | C110(0.89); C155(0.92); C160(0.93) | LDD1604 | [18] |
| LDCM0401 | CL14 | HEK-293T | C110(1.03); C155(1.04); C160(1.01) | LDD1605 | [18] |
| LDCM0402 | CL15 | HEK-293T | C110(0.89); C155(0.96); C160(0.89) | LDD1606 | [18] |
| LDCM0403 | CL16 | HEK-293T | C110(1.03); C155(1.04); C160(0.96) | LDD1607 | [18] |
| LDCM0404 | CL17 | HEK-293T | C110(1.15); C155(1.08); C160(0.98) | LDD1608 | [18] |
| LDCM0405 | CL18 | HEK-293T | C110(1.16); C155(1.02); C160(0.96) | LDD1609 | [18] |
| LDCM0406 | CL19 | HEK-293T | C110(0.89); C155(0.98); C160(1.19) | LDD1610 | [18] |
| LDCM0407 | CL2 | HEK-293T | C110(1.18); C155(1.01); C160(0.90) | LDD1611 | [18] |
| LDCM0408 | CL20 | HEK-293T | C110(1.12); C155(1.26); C160(1.16) | LDD1612 | [18] |
| LDCM0409 | CL21 | HEK-293T | C110(1.06); C155(1.17); C160(1.00) | LDD1613 | [18] |
| LDCM0410 | CL22 | HEK-293T | C110(1.16); C155(1.07); C160(1.14) | LDD1614 | [18] |
| LDCM0411 | CL23 | HEK-293T | C110(0.97); C160(1.11) | LDD1615 | [18] |
| LDCM0412 | CL24 | HEK-293T | C110(0.94); C155(1.17); C160(1.35); C155(1.21) | LDD1616 | [18] |
| LDCM0413 | CL25 | HEK-293T | C110(0.96); C155(1.02); C160(0.83) | LDD1617 | [18] |
| LDCM0414 | CL26 | HEK-293T | C110(1.02); C155(0.91); C160(0.99) | LDD1618 | [18] |
| LDCM0415 | CL27 | HEK-293T | C110(0.96); C155(0.91); C160(0.98) | LDD1619 | [18] |
| LDCM0416 | CL28 | HEK-293T | C110(1.05); C155(1.06); C160(0.96) | LDD1620 | [18] |
| LDCM0417 | CL29 | HEK-293T | C110(1.07); C155(0.89); C160(0.99) | LDD1621 | [18] |
| LDCM0418 | CL3 | HEK-293T | C110(0.85); C155(0.91); C160(0.97) | LDD1622 | [18] |
| LDCM0419 | CL30 | HEK-293T | C110(1.05); C155(0.94); C160(0.97) | LDD1623 | [18] |
| LDCM0420 | CL31 | HEK-293T | C110(0.99); C155(1.06); C160(1.13) | LDD1624 | [18] |
| LDCM0421 | CL32 | HEK-293T | C110(1.05); C155(1.20); C160(1.19) | LDD1625 | [18] |
| LDCM0422 | CL33 | HEK-293T | C110(1.09); C155(1.11); C160(0.93) | LDD1626 | [18] |
| LDCM0423 | CL34 | HEK-293T | C110(1.16); C155(0.99); C160(0.97) | LDD1627 | [18] |
| LDCM0424 | CL35 | HEK-293T | C110(1.07); C160(1.00) | LDD1628 | [18] |
| LDCM0425 | CL36 | HEK-293T | C110(0.92); C155(1.08); C160(1.17); C155(1.03) | LDD1629 | [18] |
| LDCM0426 | CL37 | HEK-293T | C110(1.07); C155(0.91); C160(0.89) | LDD1630 | [18] |
| LDCM0428 | CL39 | HEK-293T | C110(1.02); C155(0.89); C160(0.90) | LDD1632 | [18] |
| LDCM0429 | CL4 | HEK-293T | C110(1.02); C155(0.99); C160(0.99) | LDD1633 | [18] |
| LDCM0430 | CL40 | HEK-293T | C110(1.12); C155(1.04); C160(1.01) | LDD1634 | [18] |
| LDCM0431 | CL41 | HEK-293T | C110(1.05); C155(1.07); C160(0.97) | LDD1635 | [18] |
| LDCM0432 | CL42 | HEK-293T | C110(1.05); C155(1.01); C160(0.92) | LDD1636 | [18] |
| LDCM0433 | CL43 | HEK-293T | C110(0.80); C155(1.00); C160(1.09) | LDD1637 | [18] |
| LDCM0434 | CL44 | HEK-293T | C110(1.11); C155(1.12); C160(1.21) | LDD1638 | [18] |
| LDCM0435 | CL45 | HEK-293T | C110(1.13); C155(1.14); C160(0.90) | LDD1639 | [18] |
| LDCM0436 | CL46 | HEK-293T | C110(1.12); C155(1.08); C160(1.11) | LDD1640 | [18] |
| LDCM0437 | CL47 | HEK-293T | C110(1.15); C160(1.11) | LDD1641 | [18] |
| LDCM0438 | CL48 | HEK-293T | C110(0.92); C155(1.18); C160(1.29); C155(1.21) | LDD1642 | [18] |
| LDCM0439 | CL49 | HEK-293T | C110(0.98); C155(0.97); C160(0.88) | LDD1643 | [18] |
| LDCM0440 | CL5 | HEK-293T | C110(1.34); C155(1.06); C160(1.11) | LDD1644 | [18] |
| LDCM0441 | CL50 | HEK-293T | C110(1.18); C155(1.00); C160(0.95) | LDD1645 | [18] |
| LDCM0443 | CL52 | HEK-293T | C110(1.19); C155(1.02); C160(0.94) | LDD1646 | [18] |
| LDCM0444 | CL53 | HEK-293T | C110(0.77); C155(1.09); C160(0.90) | LDD1647 | [18] |
| LDCM0445 | CL54 | HEK-293T | C110(1.14); C155(1.13); C160(0.90) | LDD1648 | [18] |
| LDCM0446 | CL55 | HEK-293T | C110(0.96); C155(1.01); C160(0.92) | LDD1649 | [18] |
| LDCM0447 | CL56 | HEK-293T | C110(1.13); C155(1.22); C160(1.03) | LDD1650 | [18] |
| LDCM0448 | CL57 | HEK-293T | C110(1.13); C155(1.07); C160(0.85) | LDD1651 | [18] |
| LDCM0449 | CL58 | HEK-293T | C110(1.01); C155(1.10); C160(0.95) | LDD1652 | [18] |
| LDCM0450 | CL59 | HEK-293T | C110(1.11); C160(1.00) | LDD1653 | [18] |
| LDCM0451 | CL6 | HEK-293T | C110(1.01); C155(1.02); C160(0.94) | LDD1654 | [18] |
| LDCM0452 | CL60 | HEK-293T | C110(1.05); C155(1.19); C160(1.23); C155(1.00) | LDD1655 | [18] |
| LDCM0453 | CL61 | HEK-293T | C110(0.85); C155(0.99); C160(0.97) | LDD1656 | [18] |
| LDCM0454 | CL62 | HEK-293T | C110(1.04); C155(0.79); C160(0.96) | LDD1657 | [18] |
| LDCM0455 | CL63 | HEK-293T | C110(0.80); C155(0.92); C160(0.97) | LDD1658 | [18] |
| LDCM0456 | CL64 | HEK-293T | C110(1.03); C155(1.04); C160(0.89) | LDD1659 | [18] |
| LDCM0457 | CL65 | HEK-293T | C110(1.07); C155(1.15); C160(1.03) | LDD1660 | [18] |
| LDCM0458 | CL66 | HEK-293T | C110(1.08); C155(1.11); C160(0.94) | LDD1661 | [18] |
| LDCM0459 | CL67 | HEK-293T | C110(0.93); C155(1.07); C160(0.98) | LDD1662 | [18] |
| LDCM0460 | CL68 | HEK-293T | C110(1.07); C155(1.05); C160(1.09) | LDD1663 | [18] |
| LDCM0461 | CL69 | HEK-293T | C110(1.04); C155(1.08); C160(1.12) | LDD1664 | [18] |
| LDCM0462 | CL7 | HEK-293T | C110(0.87); C155(1.08); C160(1.03) | LDD1665 | [18] |
| LDCM0463 | CL70 | HEK-293T | C110(1.02); C155(1.07); C160(0.95) | LDD1666 | [18] |
| LDCM0464 | CL71 | HEK-293T | C110(1.16); C160(1.06) | LDD1667 | [18] |
| LDCM0465 | CL72 | HEK-293T | C110(0.98); C155(1.11); C160(1.16); C155(0.97) | LDD1668 | [18] |
| LDCM0466 | CL73 | HEK-293T | C110(0.99); C155(0.96); C160(0.97) | LDD1669 | [18] |
| LDCM0467 | CL74 | HEK-293T | C110(1.09); C155(0.94); C160(1.14) | LDD1670 | [18] |
| LDCM0469 | CL76 | HEK-293T | C110(1.14); C155(1.07); C160(1.02) | LDD1672 | [18] |
| LDCM0470 | CL77 | HEK-293T | C110(0.79); C155(1.14); C160(0.94) | LDD1673 | [18] |
| LDCM0471 | CL78 | HEK-293T | C110(1.04); C155(1.04); C160(0.94) | LDD1674 | [18] |
| LDCM0472 | CL79 | HEK-293T | C110(0.97); C155(0.99); C160(1.05) | LDD1675 | [18] |
| LDCM0473 | CL8 | HEK-293T | C110(1.17); C155(1.06); C160(0.83) | LDD1676 | [18] |
| LDCM0474 | CL80 | HEK-293T | C110(1.21); C155(1.07); C160(1.19) | LDD1677 | [18] |
| LDCM0475 | CL81 | HEK-293T | C110(0.99); C155(1.12); C160(1.11) | LDD1678 | [18] |
| LDCM0476 | CL82 | HEK-293T | C110(1.06); C155(1.01); C160(0.98) | LDD1679 | [18] |
| LDCM0477 | CL83 | HEK-293T | C110(1.07); C160(1.03) | LDD1680 | [18] |
| LDCM0478 | CL84 | HEK-293T | C110(0.94); C155(1.25); C160(1.09); C155(0.93) | LDD1681 | [18] |
| LDCM0479 | CL85 | HEK-293T | C110(0.94); C155(1.00); C160(1.00) | LDD1682 | [18] |
| LDCM0480 | CL86 | HEK-293T | C110(1.06); C155(0.97); C160(0.91) | LDD1683 | [18] |
| LDCM0481 | CL87 | HEK-293T | C110(1.20); C155(0.97); C160(0.98) | LDD1684 | [18] |
| LDCM0482 | CL88 | HEK-293T | C110(1.04); C155(1.02); C160(1.04) | LDD1685 | [18] |
| LDCM0483 | CL89 | HEK-293T | C110(1.00); C155(1.08); C160(1.06) | LDD1686 | [18] |
| LDCM0484 | CL9 | HEK-293T | C110(1.03); C155(1.15); C160(1.08) | LDD1687 | [18] |
| LDCM0485 | CL90 | HEK-293T | C110(1.22); C155(1.25); C160(0.79) | LDD1688 | [18] |
| LDCM0486 | CL91 | HEK-293T | C110(0.91); C155(0.93); C160(0.91) | LDD1689 | [18] |
| LDCM0487 | CL92 | HEK-293T | C110(1.10); C155(1.11); C160(1.12) | LDD1690 | [18] |
| LDCM0488 | CL93 | HEK-293T | C110(0.95); C155(1.13); C160(1.06) | LDD1691 | [18] |
| LDCM0489 | CL94 | HEK-293T | C110(1.13); C155(1.12); C160(0.97) | LDD1692 | [18] |
| LDCM0490 | CL95 | HEK-293T | C110(0.99); C160(0.91) | LDD1693 | [18] |
| LDCM0491 | CL96 | HEK-293T | C110(0.79); C155(1.18); C160(1.31); C155(1.07) | LDD1694 | [18] |
| LDCM0492 | CL97 | HEK-293T | C110(0.89); C155(0.99); C160(0.95) | LDD1695 | [18] |
| LDCM0493 | CL98 | HEK-293T | C110(1.22); C155(1.07); C160(0.91) | LDD1696 | [18] |
| LDCM0494 | CL99 | HEK-293T | C110(0.97); C155(0.91); C160(0.92) | LDD1697 | [18] |
| LDCM0028 | Dobutamine | HEK-293T | 4.82 | LDD0180 | [5] |
| LDCM0027 | Dopamine | HEK-293T | 5.31 | LDD0179 | [5] |
| LDCM0495 | E2913 | HEK-293T | C110(1.09); C155(0.75); C160(0.92) | LDD1698 | [18] |
| LDCM0625 | F8 | Ramos | C110(1.05); 0.42 | LDD2187 | [19] |
| LDCM0572 | Fragment10 | Ramos | C110(0.87); 1.35 | LDD2189 | [19] |
| LDCM0573 | Fragment11 | Ramos | C110(0.14); 0.63 | LDD2190 | [19] |
| LDCM0574 | Fragment12 | Ramos | C110(0.78); 2.65 | LDD2191 | [19] |
| LDCM0575 | Fragment13 | Ramos | C110(0.82) | LDD2192 | [19] |
| LDCM0576 | Fragment14 | Ramos | C110(1.61); 1.81 | LDD2193 | [19] |
| LDCM0579 | Fragment20 | Ramos | C110(1.12); 1.44 | LDD2194 | [19] |
| LDCM0580 | Fragment21 | Ramos | C110(1.29); 1.20 | LDD2195 | [19] |
| LDCM0582 | Fragment23 | Ramos | C110(0.40); 1.11 | LDD2196 | [19] |
| LDCM0578 | Fragment27 | Ramos | C110(1.40); 0.88 | LDD2197 | [19] |
| LDCM0586 | Fragment28 | Ramos | C110(0.71); 0.72 | LDD2198 | [19] |
| LDCM0588 | Fragment30 | Ramos | C110(0.35); 1.09 | LDD2199 | [19] |
| LDCM0589 | Fragment31 | Ramos | C110(0.59); 1.09 | LDD2200 | [19] |
| LDCM0590 | Fragment32 | Ramos | C110(0.51); 0.43 | LDD2201 | [19] |
| LDCM0468 | Fragment33 | HEK-293T | C110(0.87); C155(0.96); C160(0.94) | LDD1671 | [18] |
| LDCM0596 | Fragment38 | Ramos | C110(0.81); 1.05 | LDD2203 | [19] |
| LDCM0566 | Fragment4 | Ramos | C110(1.00); 0.47 | LDD2184 | [19] |
| LDCM0427 | Fragment51 | HEK-293T | C110(1.29); C155(0.93); C160(0.88) | LDD1631 | [18] |
| LDCM0610 | Fragment52 | Ramos | C110(0.58); 0.74 | LDD2204 | [19] |
| LDCM0614 | Fragment56 | Ramos | C110(1.03); 1.89 | LDD2205 | [19] |
| LDCM0569 | Fragment7 | Ramos | C110(1.25); 1.08 | LDD2186 | [19] |
| LDCM0571 | Fragment9 | Ramos | C110(0.87) | LDD2188 | [19] |
| LDCM0022 | KB02 | HEK-293T | C155(0.82); C160(1.00); C110(1.14); C155(0.99) | LDD1492 | [18] |
| LDCM0023 | KB03 | HEK-293T | C155(1.08); C160(1.00); C110(1.11); C155(1.08) | LDD1497 | [18] |
| LDCM0024 | KB05 | COLO792 | C139(1.56) | LDD3310 | [4] |
| LDCM0032 | Oleacein | HEK-293T | 5.30 | LDD0184 | [5] |
| LDCM0014 | Panhematin | hPBMC | 4.32 | LDD0064 | [6] |
| LDCM0029 | Quercetin | HEK-293T | 7.60 | LDD0181 | [5] |
| LDCM0131 | RA190 | MM1.R | C110(1.22); C155(0.92); C160(0.92) | LDD0304 | [20] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Very long chain fatty acid elongase 4 (ELOVL4) | ELO family | Q9GZR5 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Aquaporin-6 (AQP6) | MIP/aquaporin (TC 1.A.8) family | Q13520 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Tax1-binding protein 1 (TAX1BP1) | . | Q86VP1 | |||
References
