Details of the Target
General Information of Target
| Target ID | LDTP06371 | |||||
|---|---|---|---|---|---|---|
| Target Name | GTP-binding protein Rheb (RHEB) | |||||
| Gene Name | RHEB | |||||
| Gene ID | 6009 | |||||
| Synonyms |
RHEB2; GTP-binding protein Rheb; Ras homolog enriched in brain; EC 3.6.5.- |
|||||
| 3D Structure | ||||||
| Sequence |
MPQSKSRKIAILGYRSVGKSSLTIQFVEGQFVDSYDPTIENTFTKLITVNGQEYHLQLVD
TAGQDEYSIFPQTYSIDINGYILVYSVTSIKSFEVIKVIHGKLLDMVGKVQIPIMLVGNK KDLHMERVISYEEGKALAESWNAAFLESSAKENQTAVDVFRRIILEAEKMDGAASQGKSS CSVM |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Small GTPase superfamily, Rheb family
|
|||||
| Subcellular location |
Endomembrane system
|
|||||
| Function |
Small GTPase that acts as an allosteric activator of the canonical mTORC1 complex, an evolutionarily conserved central nutrient sensor that stimulates anabolic reactions and macromolecule biosynthesis to promote cellular biomass generation and growth . In response to nutrients, growth factors or amino acids, specifically activates the protein kinase activity of MTOR, the catalytic component of the mTORC1 complex: acts by causing a conformational change that allows the alignment of residues in the active site of MTOR, thereby enhancing the phosphorylation of ribosomal protein S6 kinase (RPS6KB1 and RPS6KB2) and EIF4EBP1 (4E-BP1). RHEB is also required for localization of the TSC-TBC complex to lysosomal membranes. In response to starvation, RHEB is inactivated by the TSC-TBC complex, preventing activation of mTORC1. Has low intrinsic GTPase activity.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TH211 Probe Info |
 |
Y131(9.22) | LDD0260 | [1] | |
|
STPyne Probe Info |
 |
K120(8.90); K121(5.46) | LDD0277 | [2] | |
|
ONAyne Probe Info |
 |
K121(10.00) | LDD0275 | [2] | |
|
Jackson_1 Probe Info |
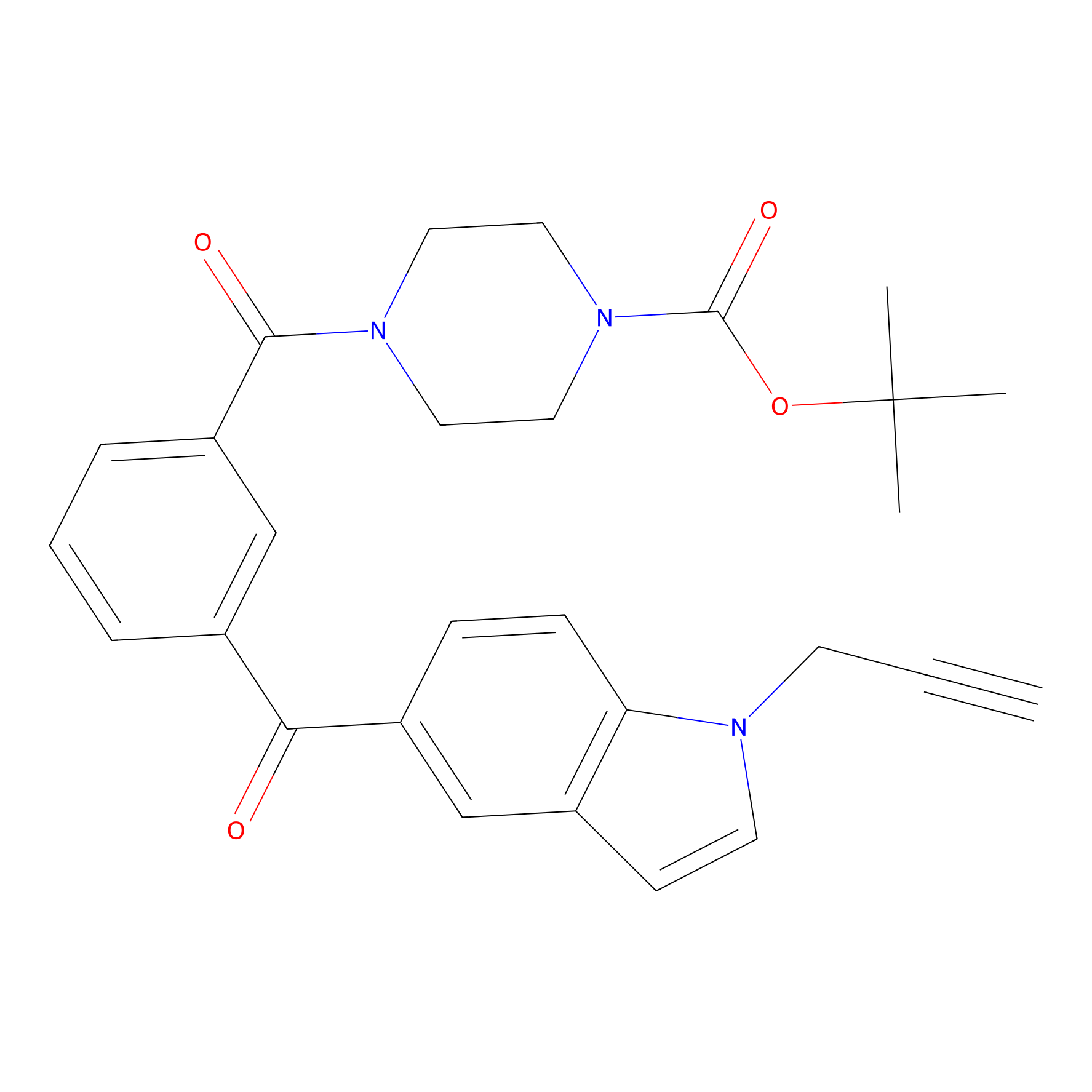 |
2.12 | LDD0120 | [3] | |
|
HHS-475 Probe Info |
 |
Y131(0.57) | LDD0265 | [4] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [5] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C094 Probe Info |
 |
28.64 | LDD1785 | [6] | |
|
C100 Probe Info |
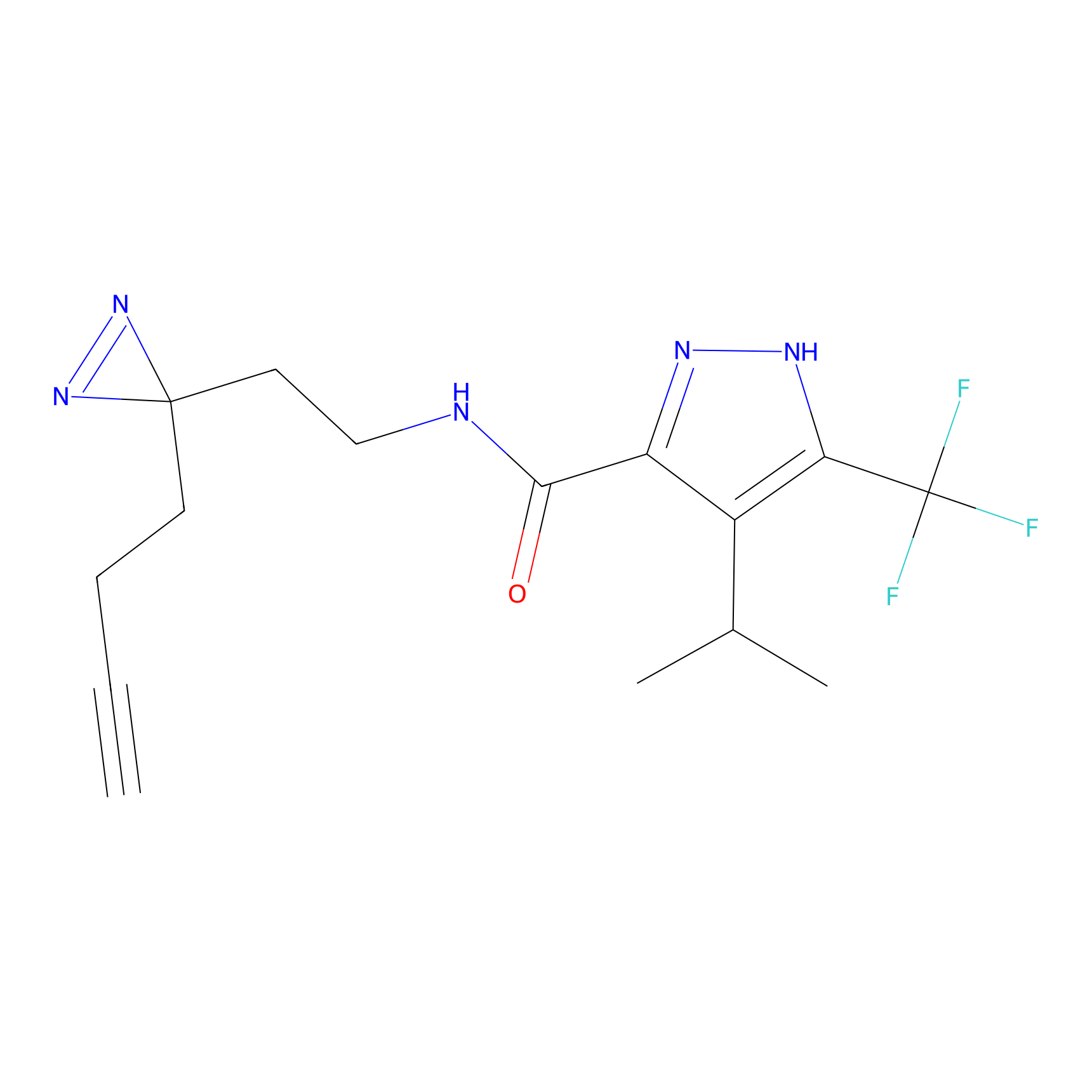 |
5.17 | LDD1789 | [6] | |
|
C153 Probe Info |
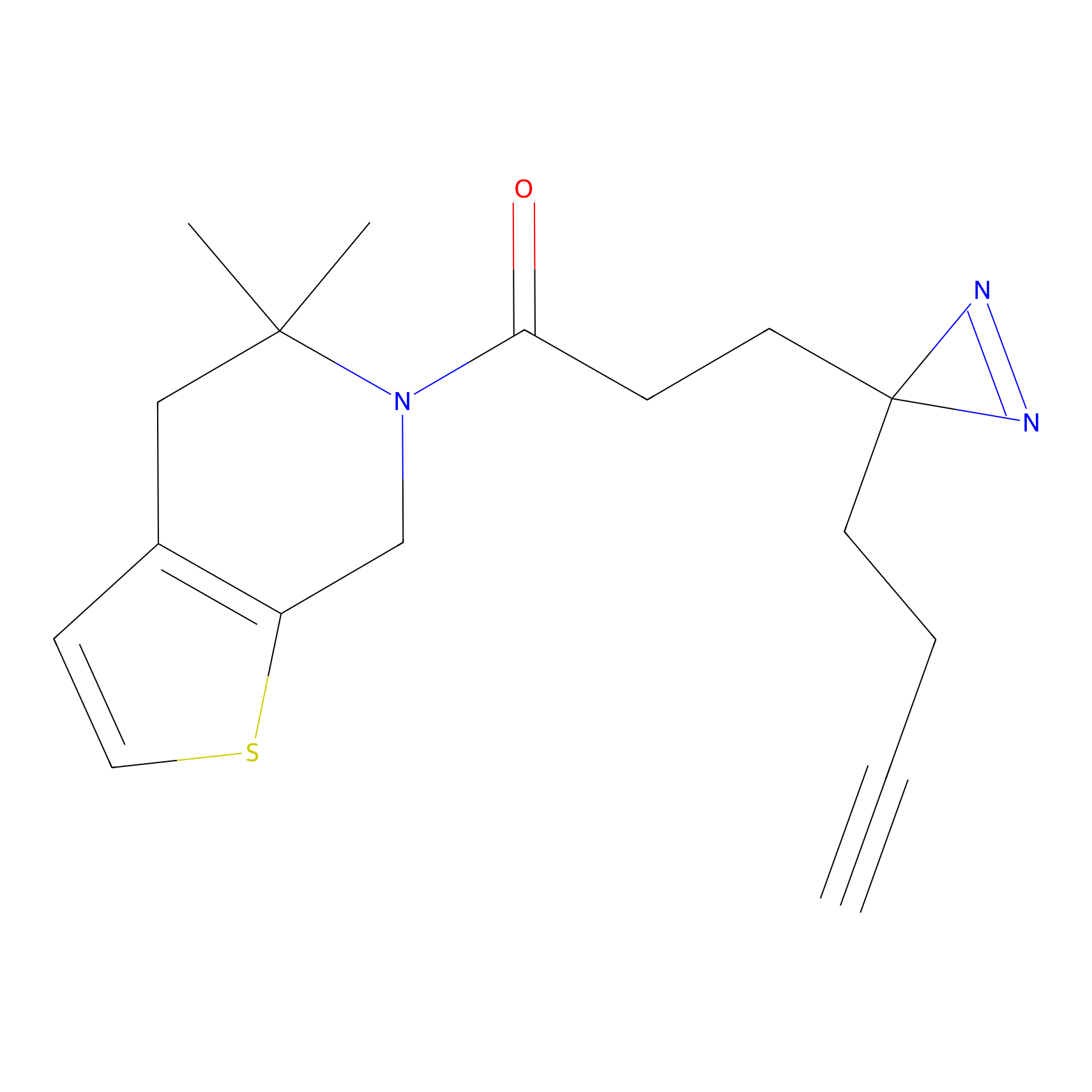 |
17.39 | LDD1834 | [6] | |
|
C158 Probe Info |
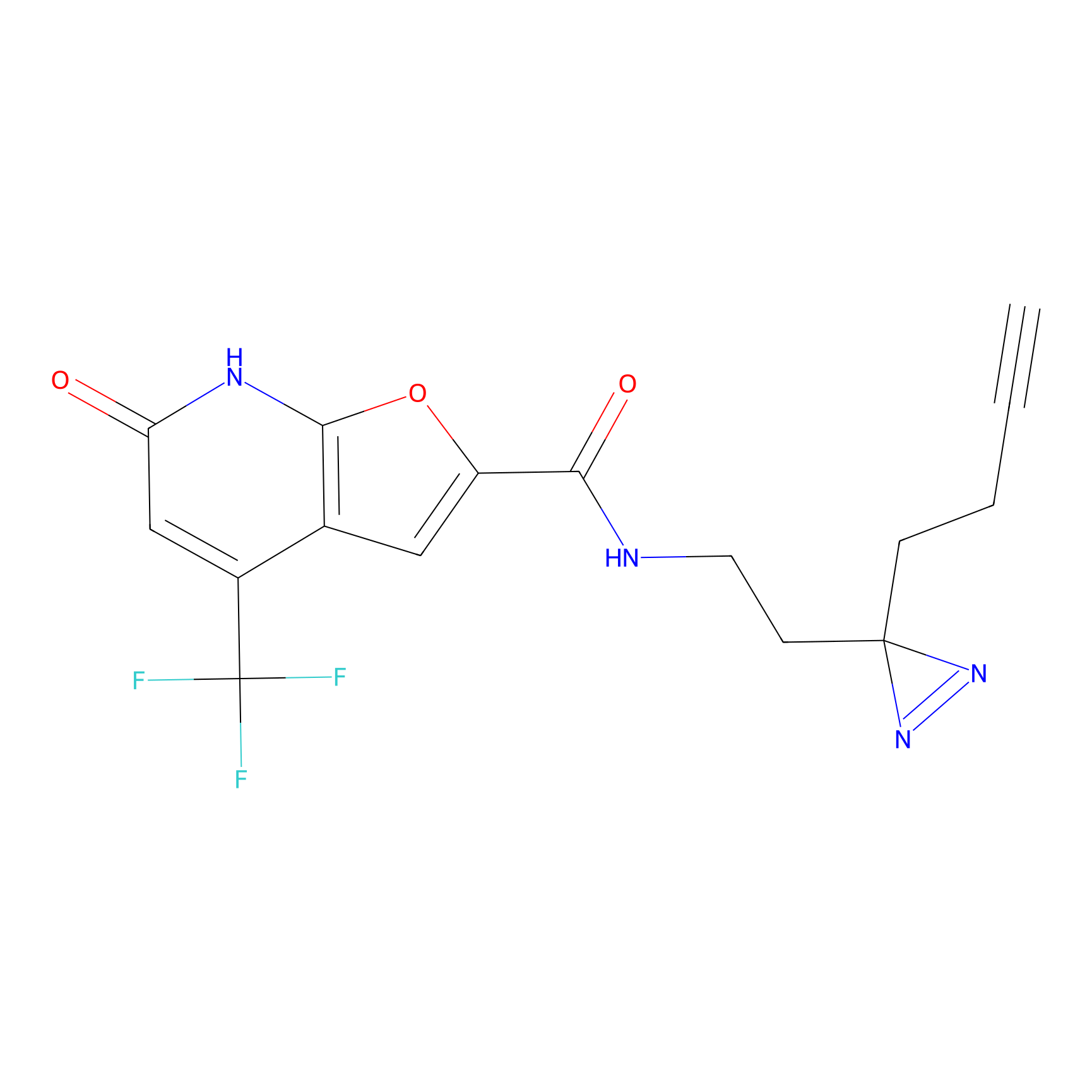 |
13.83 | LDD1838 | [6] | |
|
C232 Probe Info |
 |
37.79 | LDD1905 | [6] | |
|
C235 Probe Info |
 |
22.63 | LDD1908 | [6] | |
|
C293 Probe Info |
 |
18.25 | LDD1963 | [6] | |
|
C299 Probe Info |
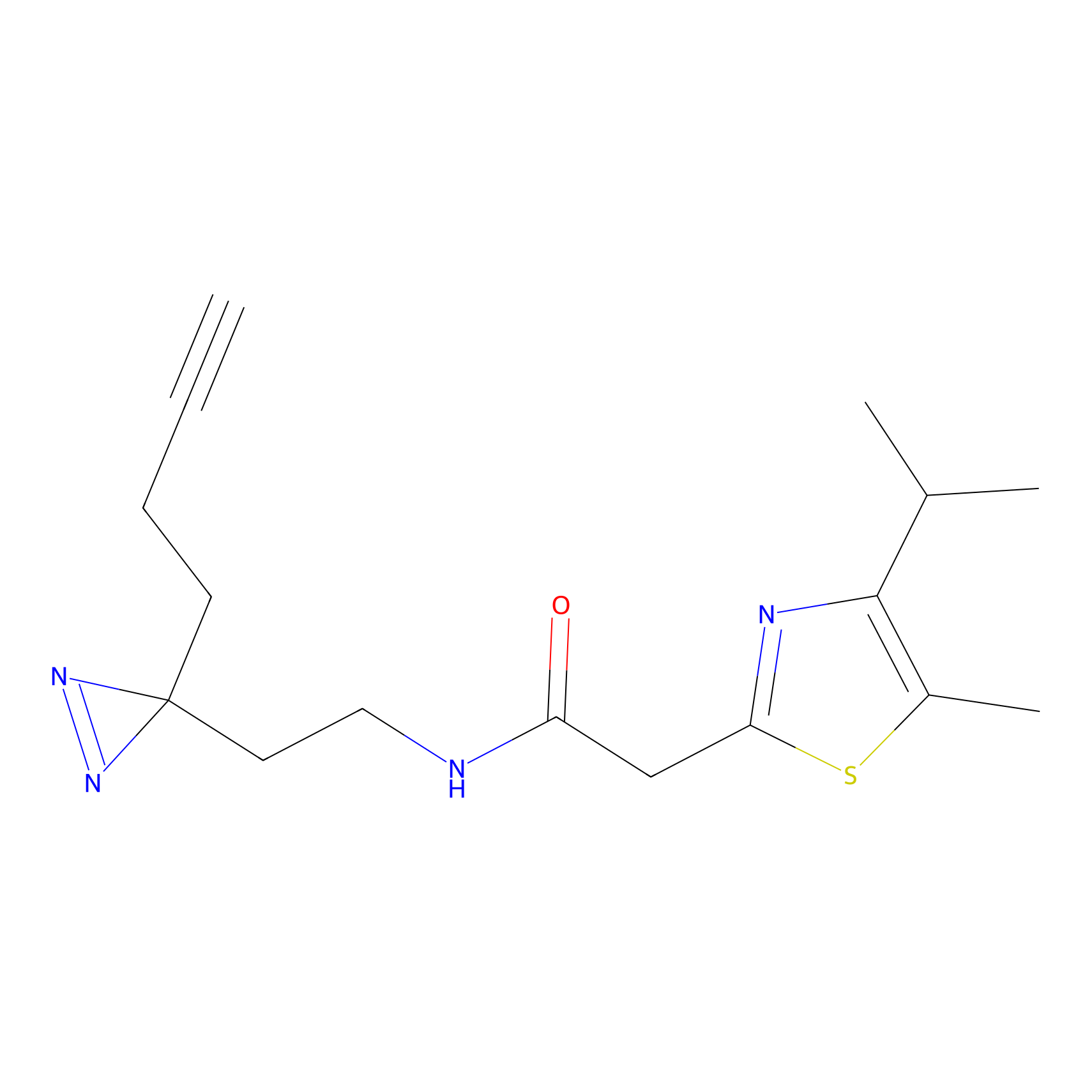 |
6.73 | LDD1968 | [6] | |
|
C305 Probe Info |
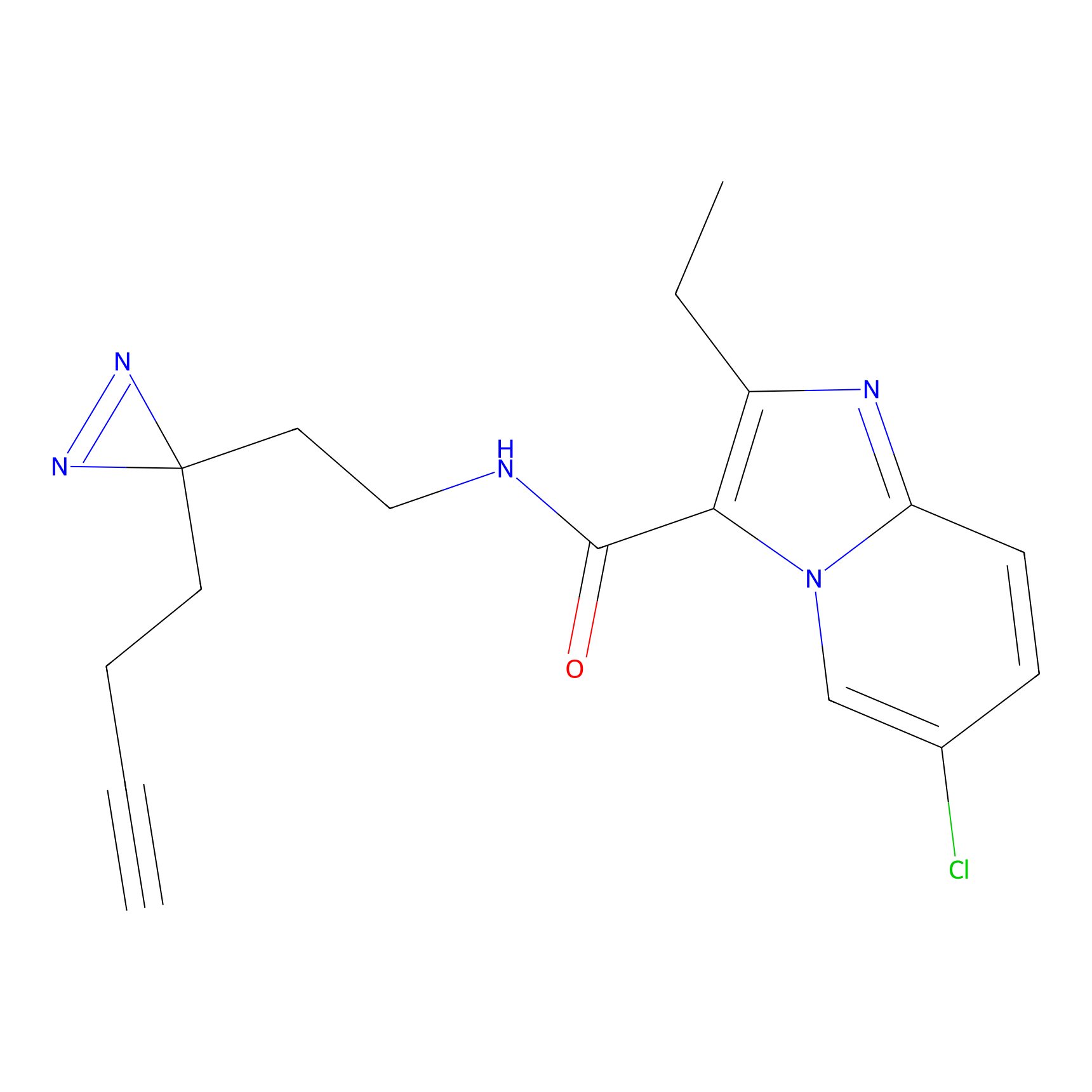 |
11.55 | LDD1974 | [6] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [7] | |
|
FFF probe3 Probe Info |
 |
20.00 | LDD0464 | [7] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0070 | Cisar_cp37 | MDA-MB-231 | 2.12 | LDD0120 | [3] |
| LDCM0191 | Compound 21 | HEK-293T | 7.09 | LDD0508 | [7] |
| LDCM0190 | Compound 34 | HEK-293T | 10.47 | LDD0510 | [7] |
| LDCM0192 | Compound 35 | HEK-293T | 6.56 | LDD0509 | [7] |
| LDCM0193 | Compound 36 | HEK-293T | 9.26 | LDD0511 | [7] |
| LDCM0117 | HHS-0201 | DM93 | Y131(0.57) | LDD0265 | [4] |
| LDCM0118 | HHS-0301 | DM93 | Y131(0.96) | LDD0266 | [4] |
| LDCM0119 | HHS-0401 | DM93 | Y131(0.89) | LDD0267 | [4] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Glutamate receptor ionotropic, NMDA 2C (GRIN2C) | Glutamate-gated ion channel family | Q14957 | |||
| Huntingtin-interacting protein 1 (HIP1) | SLA2 family | O00291 | |||
Other
References
