Details of the Target
General Information of Target
| Target ID | LDTP02869 | |||||
|---|---|---|---|---|---|---|
| Target Name | Stathmin (STMN1) | |||||
| Gene Name | STMN1 | |||||
| Gene ID | 3925 | |||||
| Synonyms |
C1orf215; LAP18; OP18; Stathmin; Leukemia-associated phosphoprotein p18; Metablastin; Oncoprotein 18; Op18; Phosphoprotein p19; pp19; Prosolin; Protein Pr22; pp17 |
|||||
| 3D Structure | ||||||
| Sequence |
MASSDIQVKELEKRASGQAFELILSPRSKESVPEFPLSPPKKKDLSLEEIQKKLEAAEER
RKSHEAEVLKQLAEKREHEKEVLQKAIEENNNFSKMAEEKLTHKMEANKENREAQMAAKL ERLREKDKHIEEVRKNKESKDPADETEAD |
|||||
| Target Type |
Clinical trial
|
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Stathmin family
|
|||||
| Subcellular location |
Cytoplasm, cytoskeleton
|
|||||
| Function |
Involved in the regulation of the microtubule (MT) filament system by destabilizing microtubules. Prevents assembly and promotes disassembly of microtubules. Phosphorylation at Ser-16 may be required for axon formation during neurogenesis. Involved in the control of the learned and innate fear.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
11.72 | LDD0402 | [1] | |
|
AZ-9 Probe Info |
 |
10.00 | LDD0393 | [2] | |
|
FBPP2 Probe Info |
 |
4.51 | LDD0318 | [3] | |
|
SAA-alkyne Probe Info |
 |
1.08 | LDD0252 | [4] | |
|
C-Sul Probe Info |
 |
16.62 | LDD0066 | [5] | |
|
ONAyne Probe Info |
 |
K53(0.00); K119(0.00); K62(0.00); K104(0.00) | LDD0273 | [6] | |
|
OPA-S-S-alkyne Probe Info |
 |
K119(1.62); K53(1.68); K128(2.80); K126(2.80) | LDD3494 | [7] | |
|
AMP probe Probe Info |
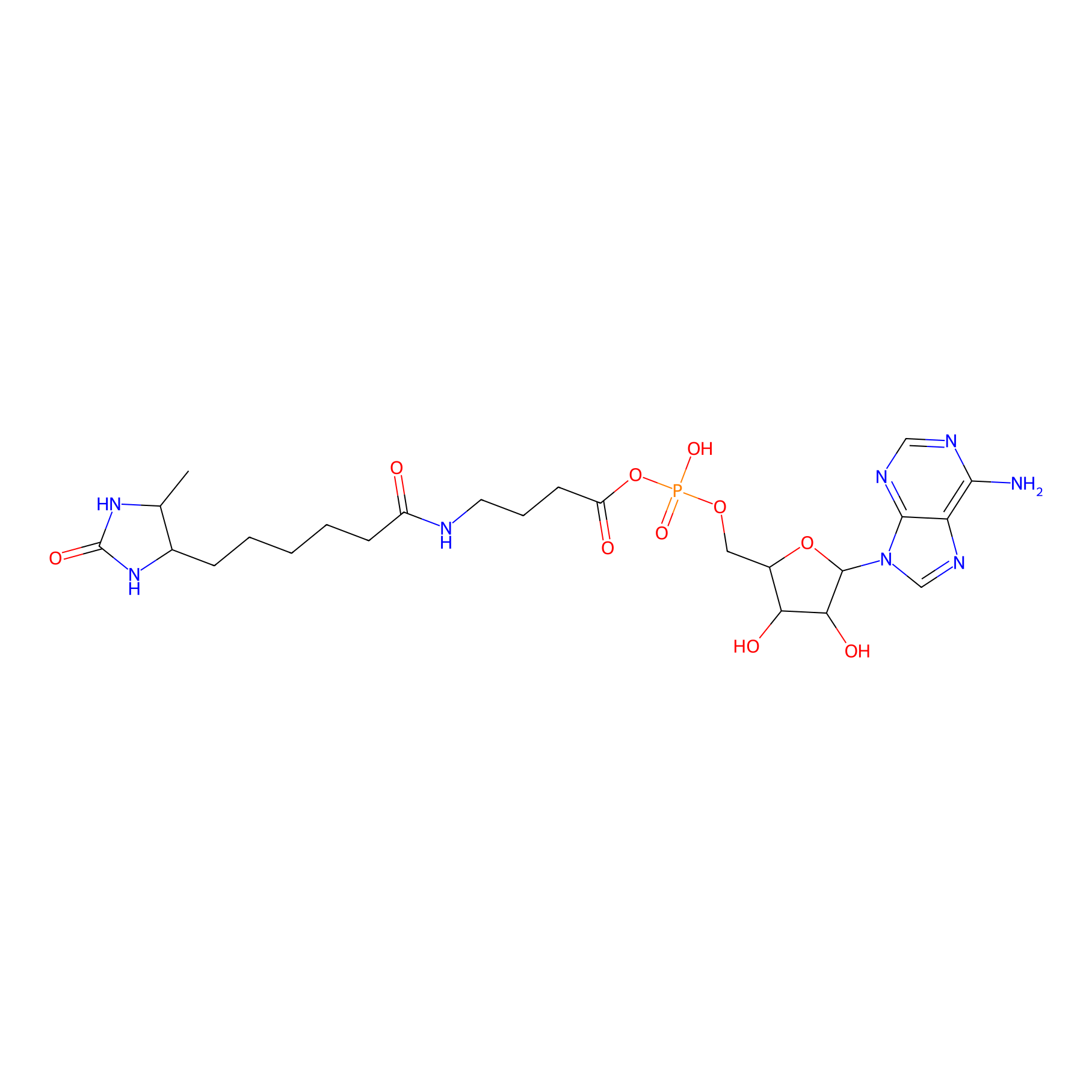 |
K53(0.00); K70(0.00) | LDD0200 | [8] | |
|
ATP probe Probe Info |
 |
K53(0.00); K70(0.00); K128(0.00); K119(0.00) | LDD0199 | [8] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [9] | |
|
NHS Probe Info |
 |
K43(0.00); K85(0.00); K100(0.00); K41(0.00) | LDD0010 | [10] | |
|
SF Probe Info |
 |
K128(0.00); K53(0.00); K29(0.00); K119(0.00) | LDD0028 | [11] | |
|
STPyne Probe Info |
 |
K85(0.00); K70(0.00); K119(0.00); K80(0.00) | LDD0009 | [10] | |
|
Acrolein Probe Info |
 |
H78(0.00); H64(0.00); K128(0.00); H129(0.00) | LDD0217 | [12] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [12] | |
|
Methacrolein Probe Info |
 |
H129(0.00); H64(0.00) | LDD0218 | [12] | |
|
AOyne Probe Info |
 |
13.70 | LDD0443 | [13] | |
|
HHS-465 Probe Info |
 |
K100(0.00); K80(0.00) | LDD2240 | [14] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C003 Probe Info |
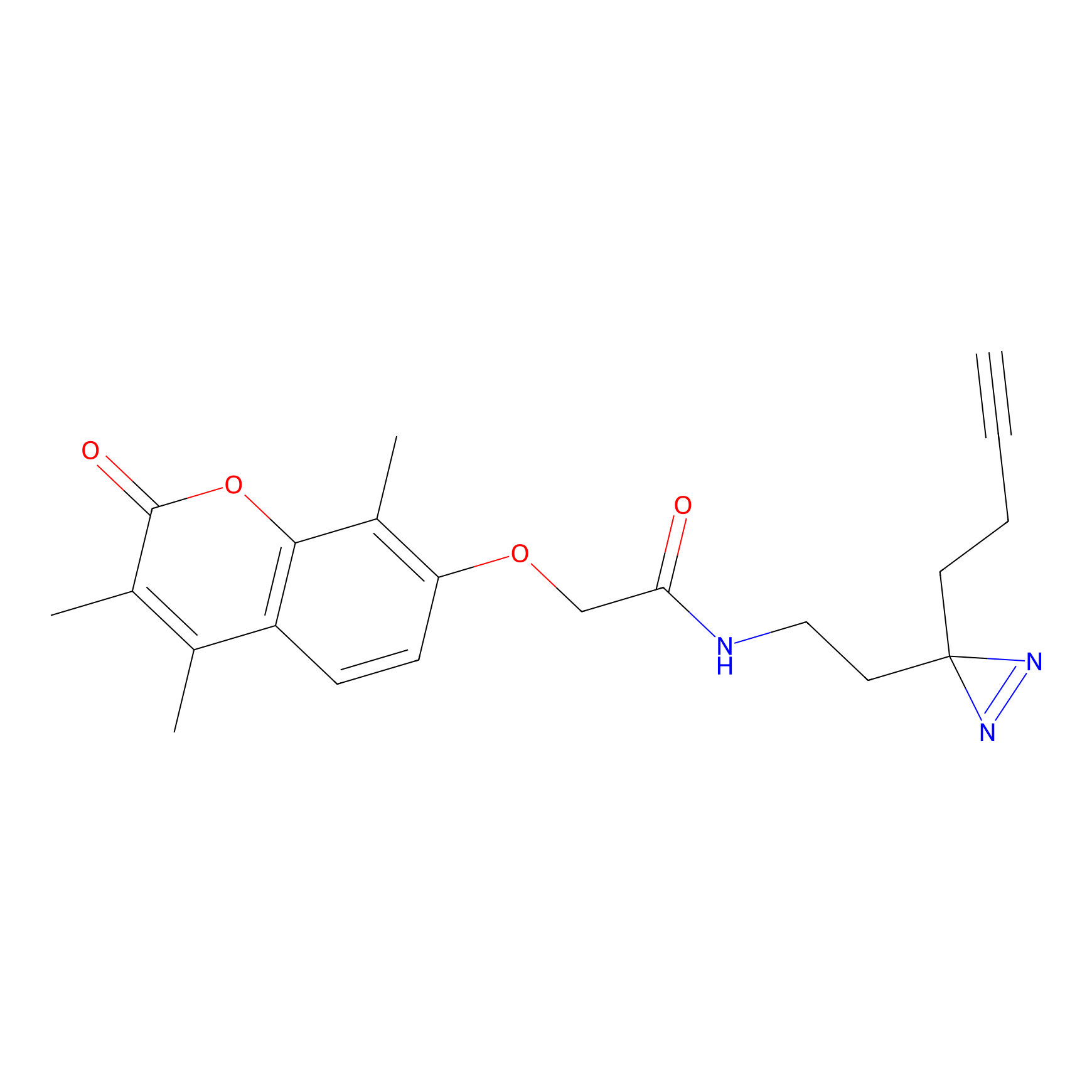 |
21.56 | LDD1713 | [15] | |
|
C052 Probe Info |
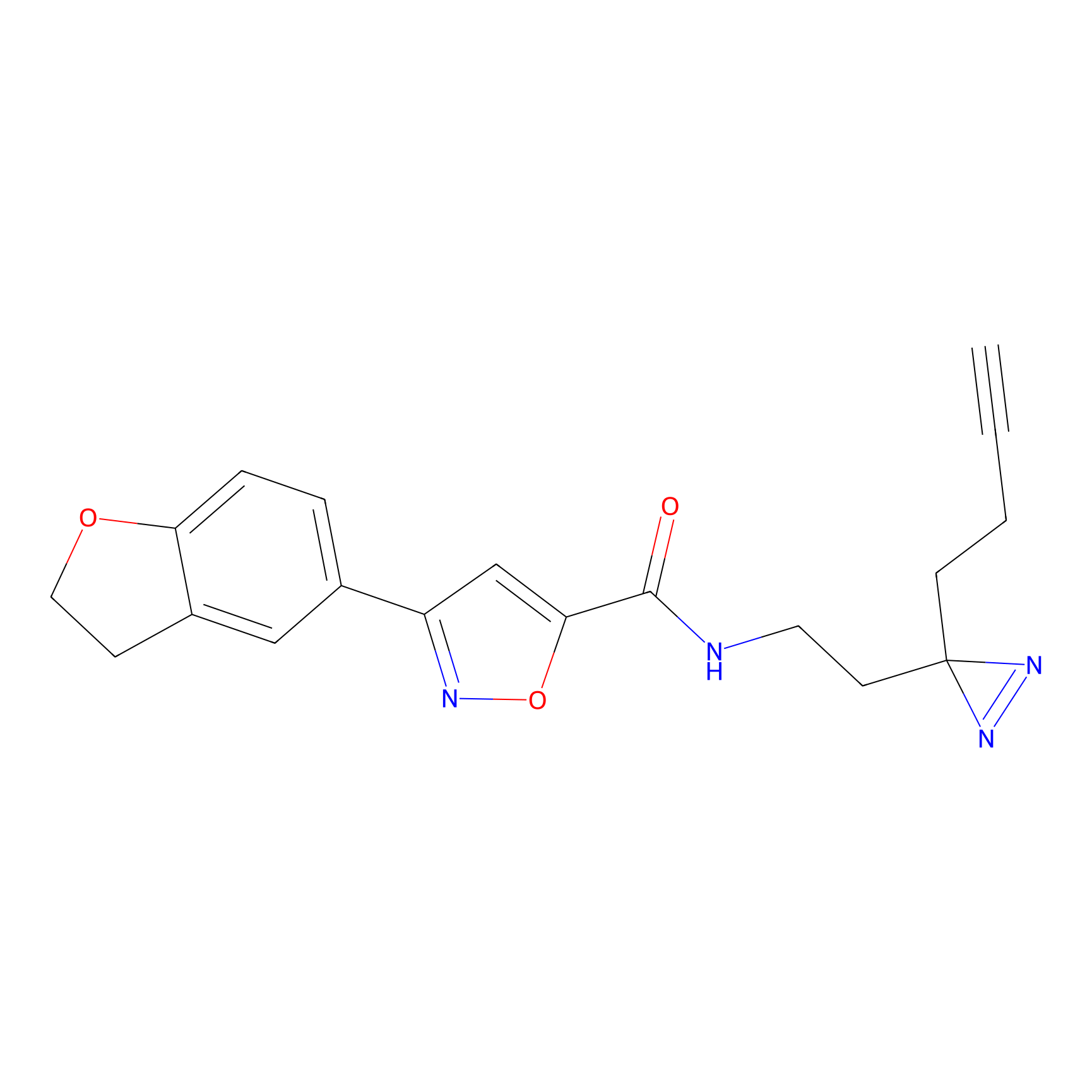 |
8.17 | LDD1750 | [15] | |
|
C187 Probe Info |
 |
16.45 | LDD1865 | [15] | |
|
C252 Probe Info |
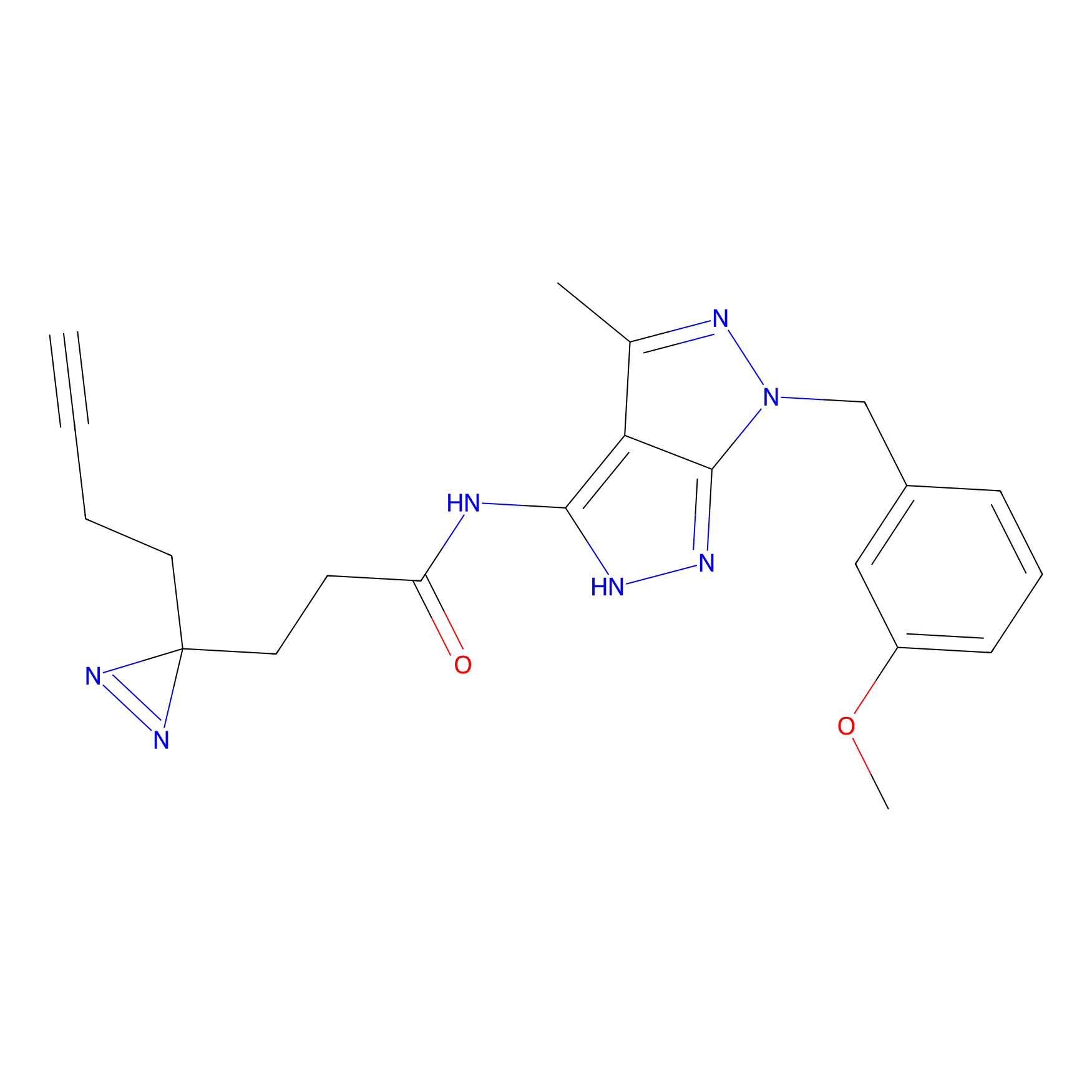 |
9.65 | LDD1925 | [15] | |
|
C314 Probe Info |
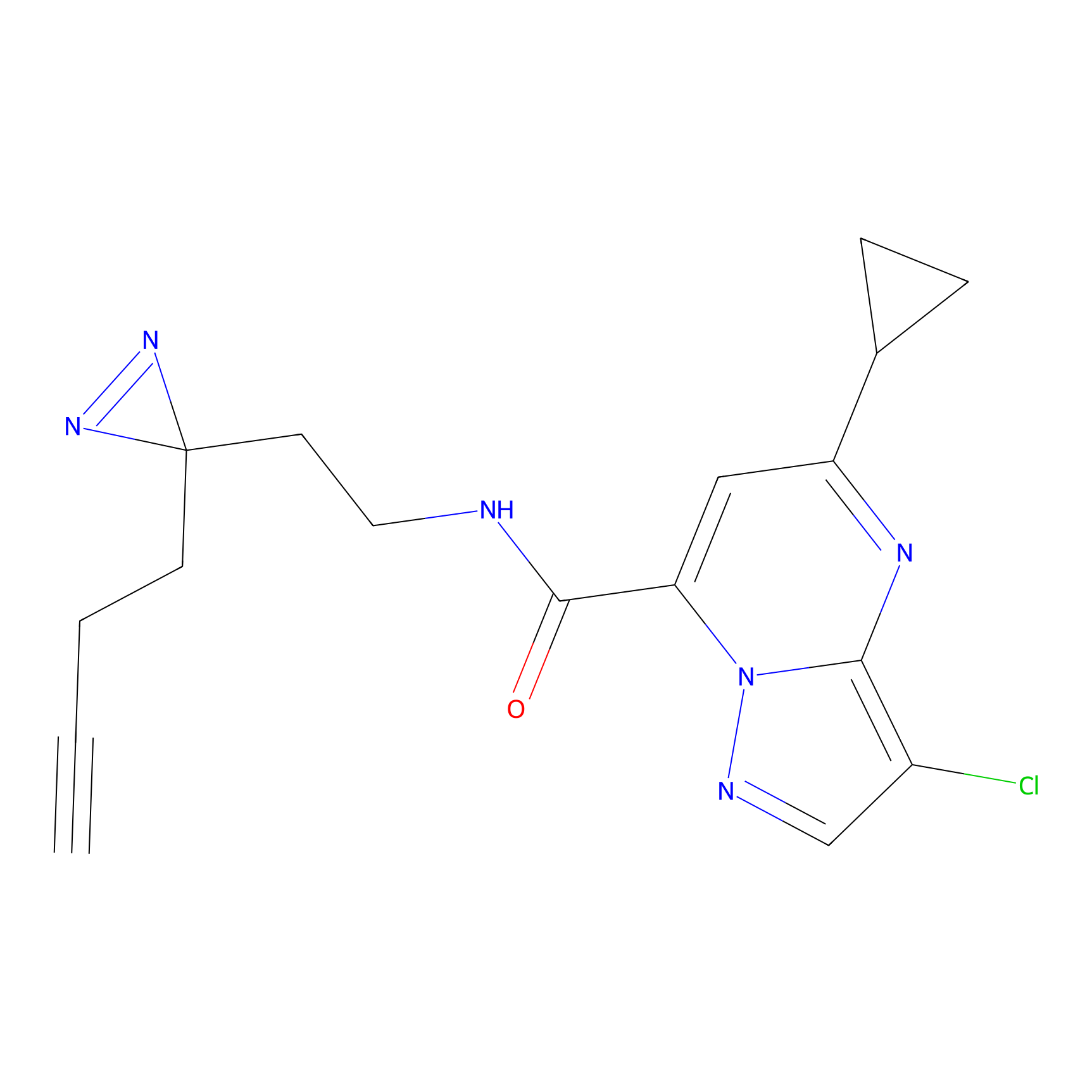 |
11.63 | LDD1981 | [15] | |
|
C354 Probe Info |
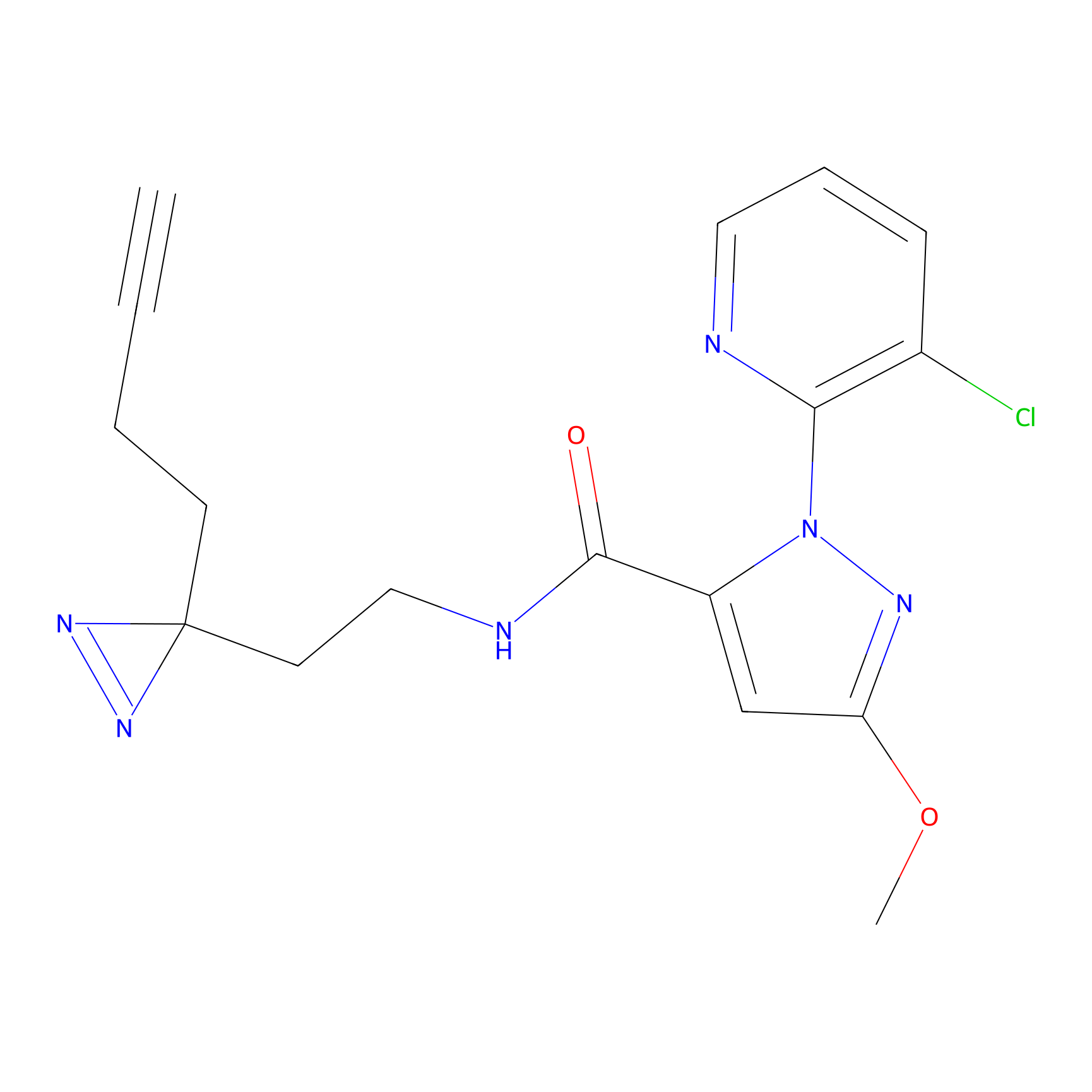 |
7.62 | LDD2015 | [15] | |
|
C397 Probe Info |
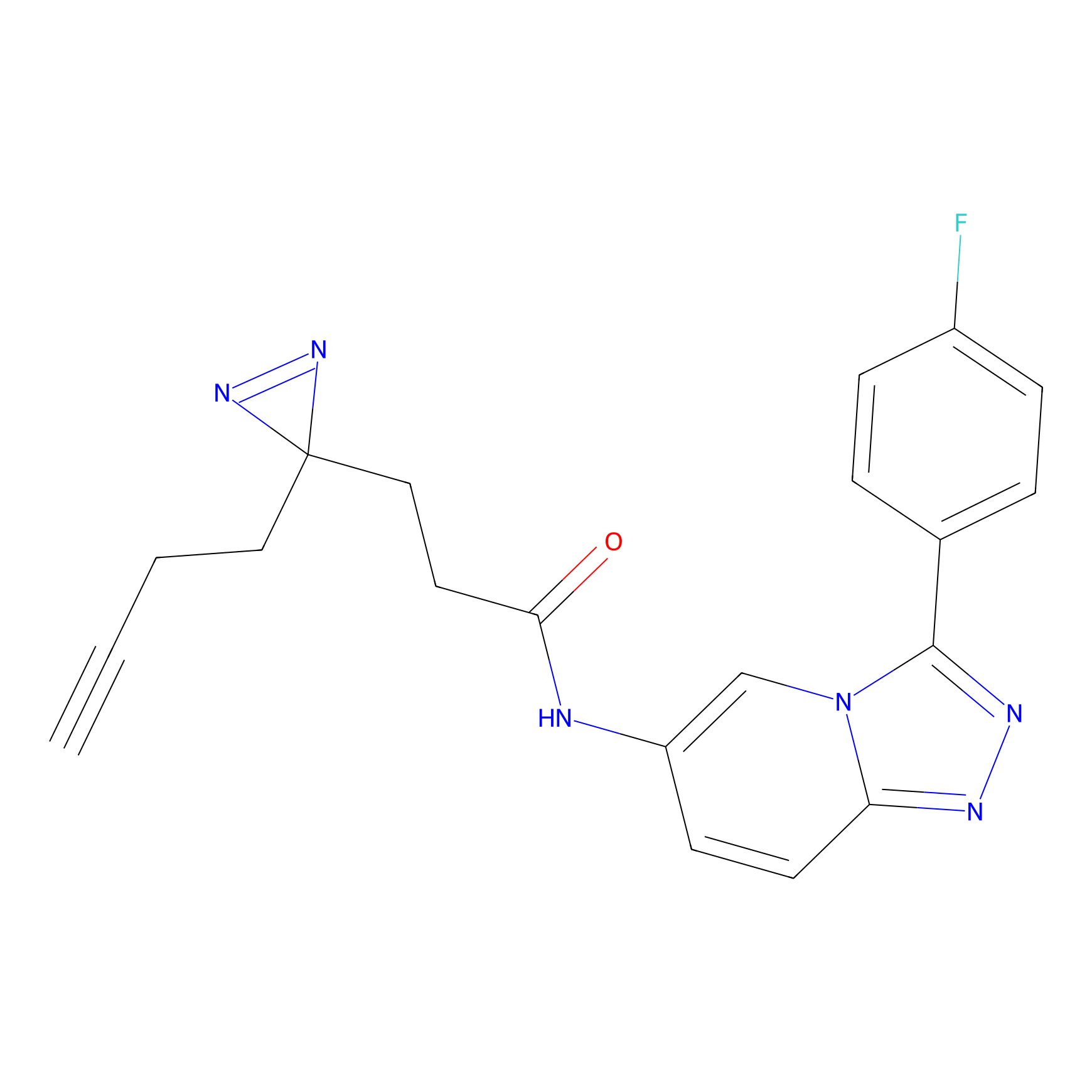 |
11.16 | LDD2056 | [15] | |
|
FFF probe12 Probe Info |
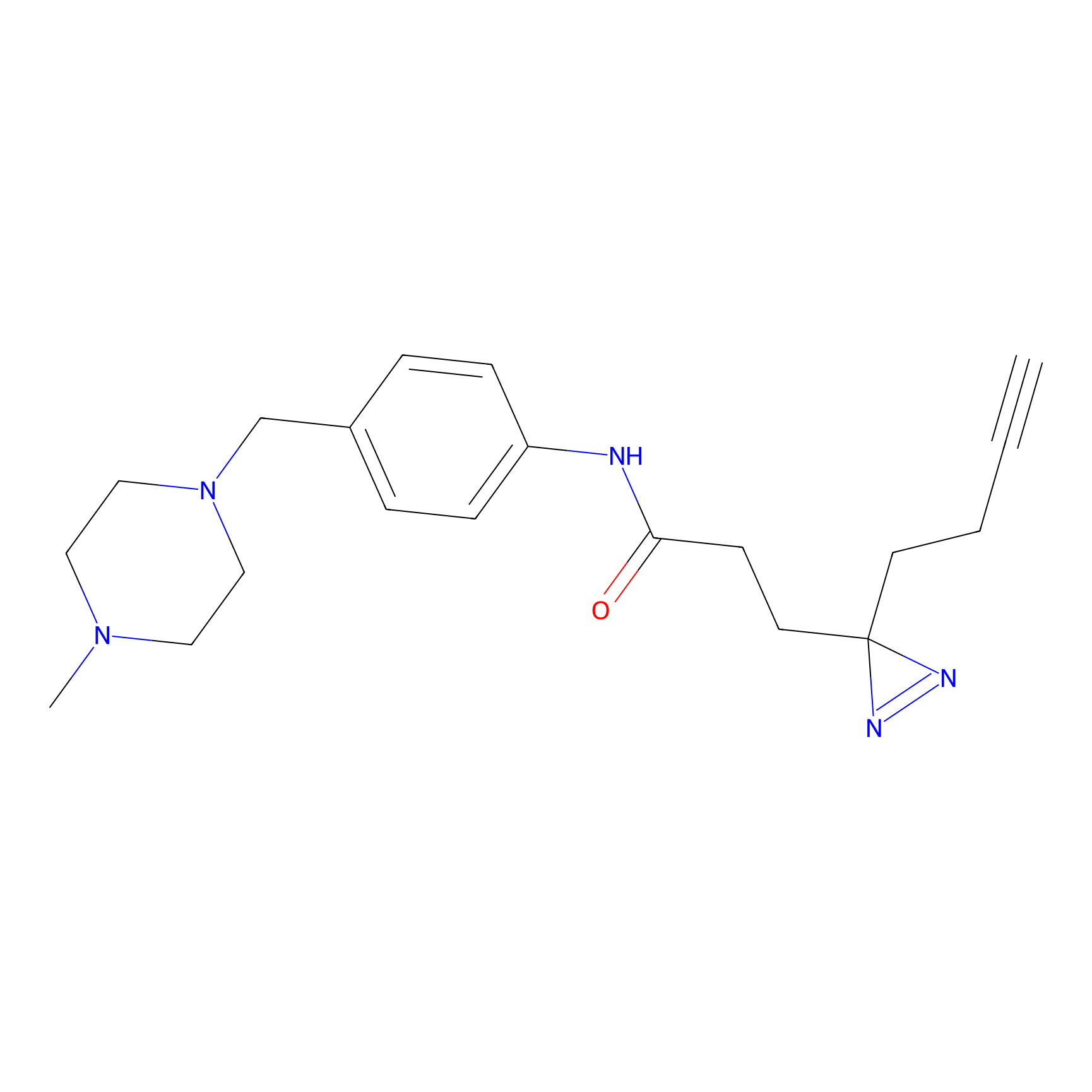 |
16.04 | LDD0473 | [16] | |
|
FFF probe13 Probe Info |
 |
12.65 | LDD0475 | [16] | |
|
FFF probe3 Probe Info |
 |
20.00 | LDD0464 | [16] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0156 | Aniline | NCI-H1299 | 12.48 | LDD0403 | [1] |
| LDCM0108 | Chloroacetamide | HeLa | H78(0.00); H129(0.00); H64(0.00) | LDD0222 | [12] |
| LDCM0107 | IAA | HeLa | H64(0.00); H129(0.00); K128(0.00) | LDD0221 | [12] |
| LDCM0109 | NEM | HeLa | H64(0.00); H129(0.00); H78(0.00); K128(0.00) | LDD0223 | [12] |
The Interaction Atlas With This Target
References
