Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TH211 Probe Info |
 |
Y431(20.00); Y611(20.00); Y115(13.30); Y525(12.77) | LDD0257 | [1] | |
|
TH214 Probe Info |
 |
Y115(20.00); Y545(18.14); Y183(15.60); Y525(11.45) | LDD0258 | [1] | |
|
TH216 Probe Info |
 |
Y115(20.00); Y183(20.00); Y545(12.76); Y611(11.98) | LDD0259 | [1] | |
|
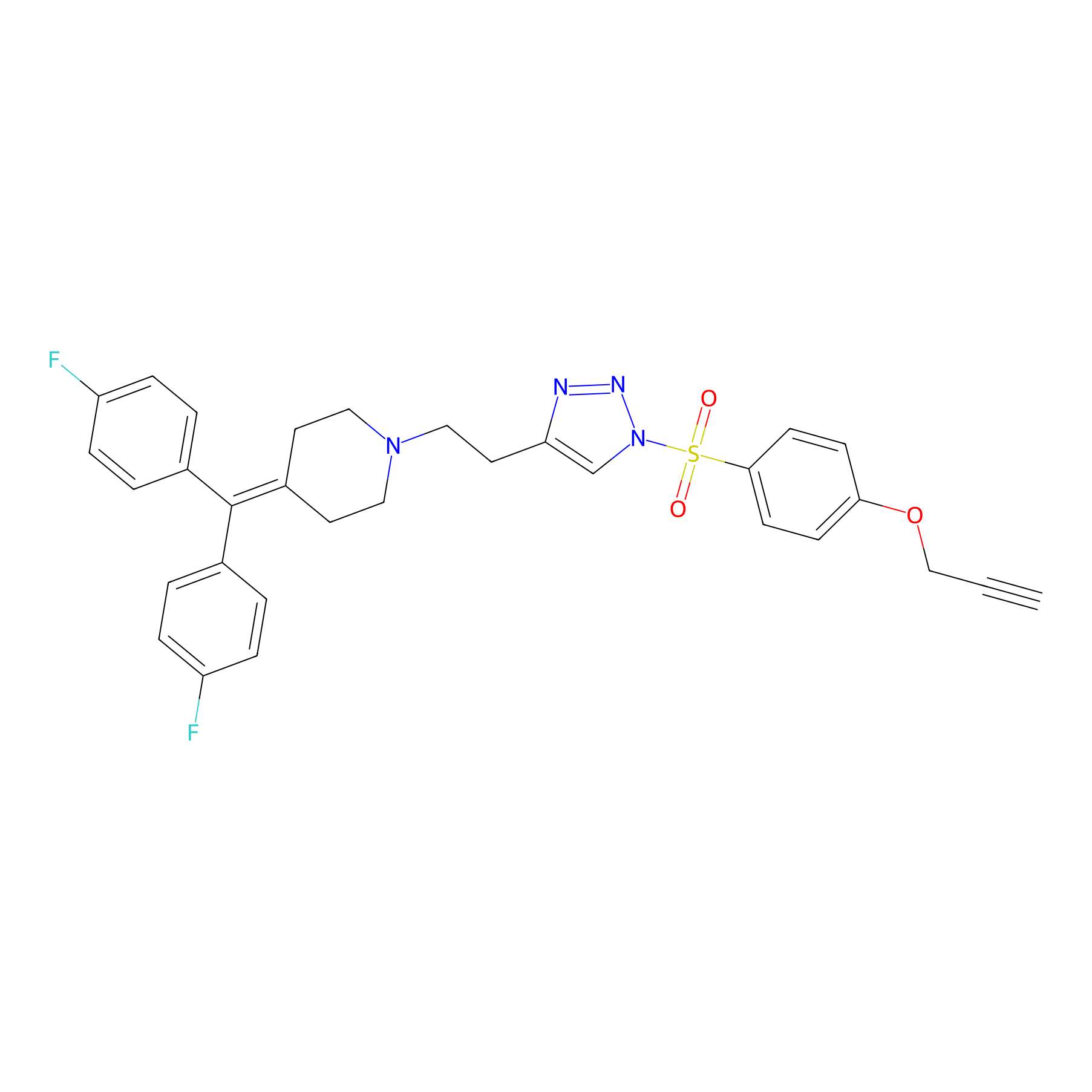
1oxF11yne Probe Info |
 |
N.A. | LDD0193 | [2] | |
|
AZ-9 Probe Info |
 |
E523(0.83); E315(1.00); E521(0.86); D160(0.70) | LDD2208 | [3] | |
|
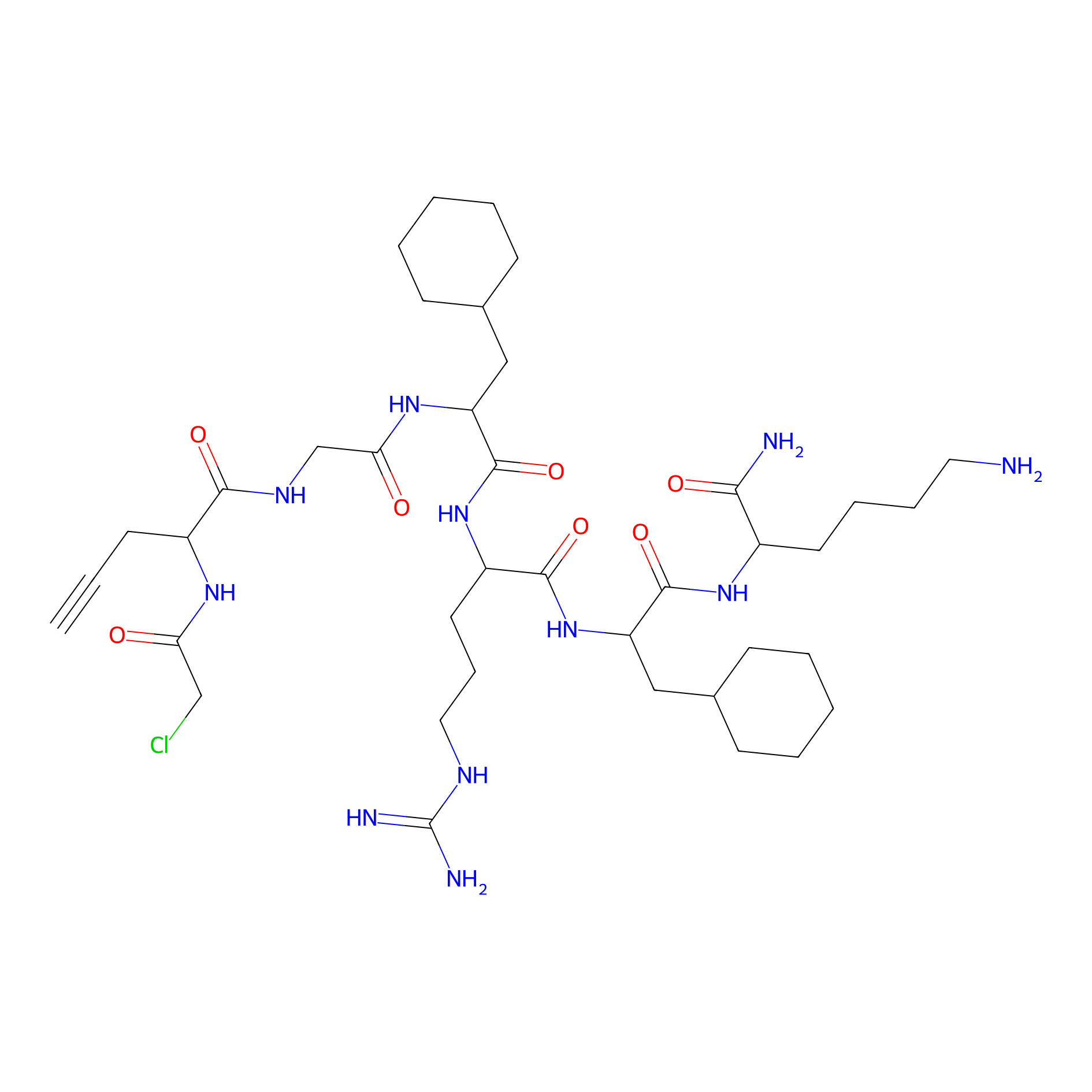
OPA-S-S-alkyne Probe Info |
 |
K595(0.41); K71(0.55); K451(1.04); K319(1.98) | LDD3494 | [4] | |
|
BTD Probe Info |
 |
C306(1.01) | LDD2112 | [5] | |
|
AHL-Pu-1 Probe Info |
 |
C603(2.17); C17(2.03) | LDD0168 | [6] | |
|
HHS-482 Probe Info |
 |
Y107(0.97); Y115(1.18); Y15(1.37); Y183(0.74) | LDD0285 | [7] | |
|
IPM Probe Info |
 |
C17(0.52) | LDD1701 | [5] | |
|
HHS-465 Probe Info |
 |
Y107(10.00); Y115(10.00); Y134(3.57); Y137(5.42) | LDD2237 | [8] | |
|
5E-2FA Probe Info |
 |
H23(0.00); H332(0.00); H227(0.00); H594(0.00) | LDD2235 | [9] | |
|
m-APA Probe Info |
 |
H23(0.00); H332(0.00); H227(0.00); H594(0.00) | LDD2231 | [9] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [10] | |
|
IA-alkyne Probe Info |
 |
C306(0.00); C574(0.00) | LDD0036 | [10] | |
|
Lodoacetamide azide Probe Info |
 |
C17(0.00); C306(0.00); C574(0.00) | LDD0037 | [10] | |
|
1c-yne Probe Info |
 |
K524(0.00); K56(0.00); K100(0.00); K328(0.00) | LDD0228 | [11] | |
|
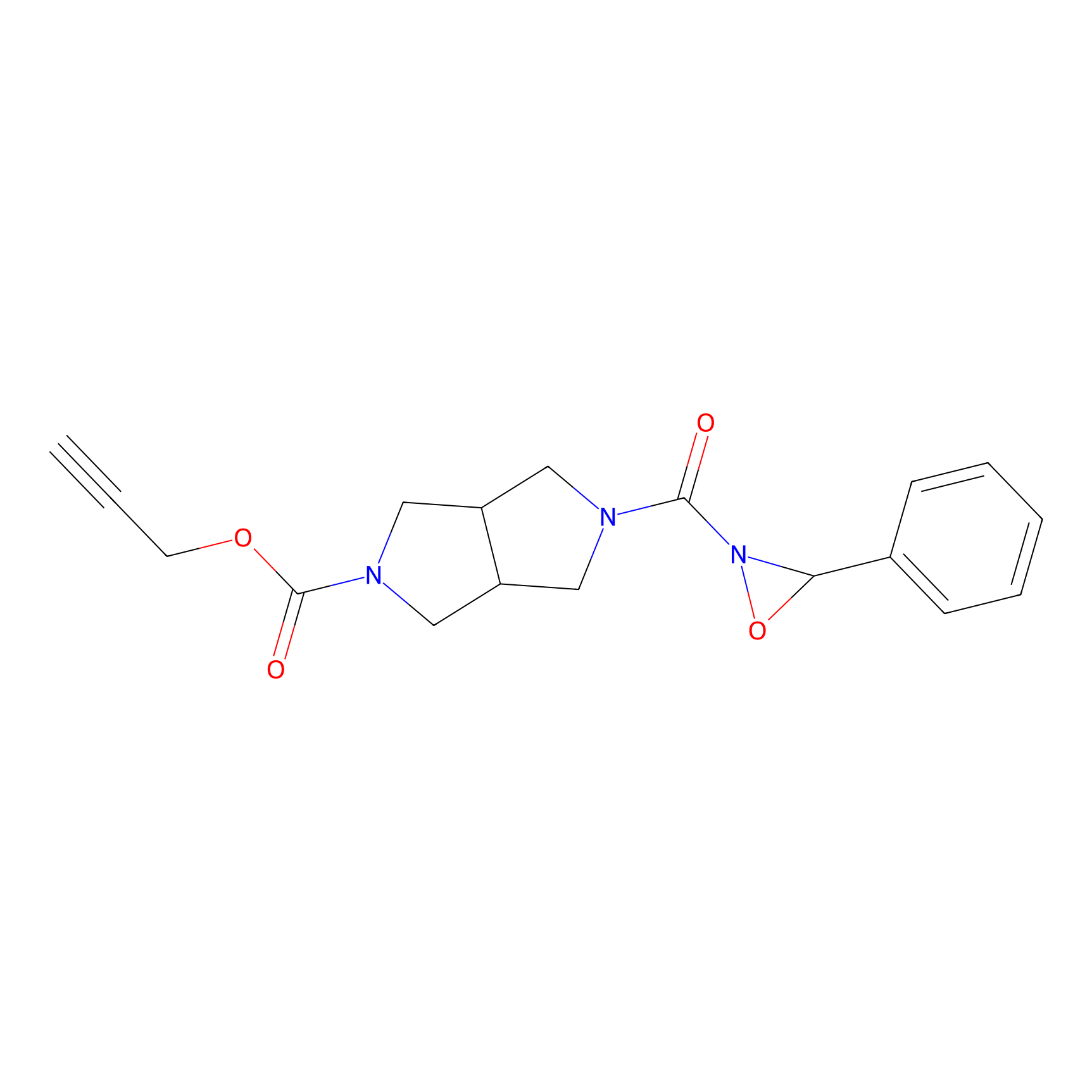
MPP-AC Probe Info |
 |
N.A. | LDD0428 | [12] | |
|
NAIA_5 Probe Info |
 |
C17(0.00); C306(0.00); C267(0.00) | LDD2223 | [13] | |
|
TPP-AC Probe Info |
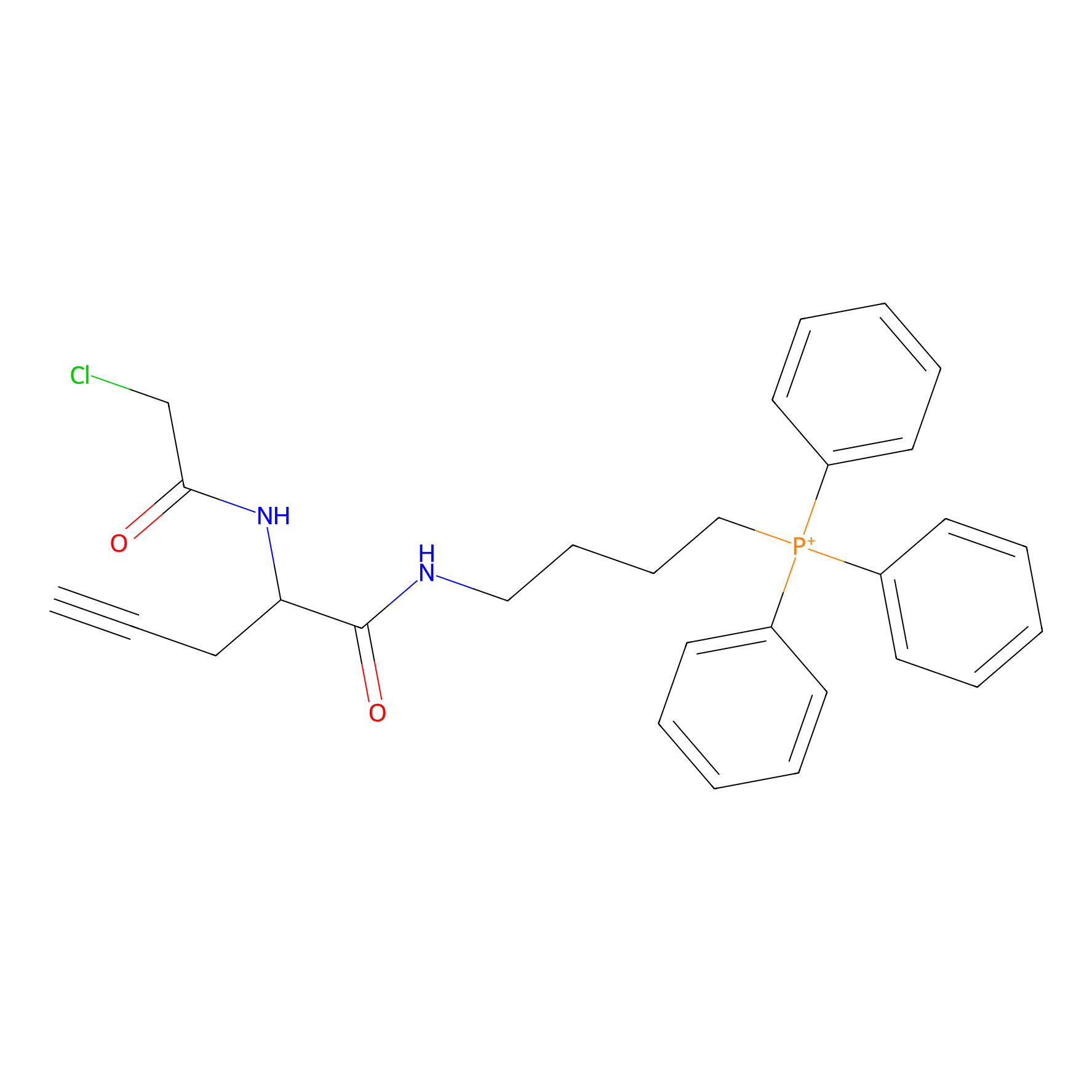 |
N.A. | LDD0427 | [12] | |
|
HHS-475 Probe Info |
 |
Y107(1.08); Y431(0.75); Y525(0.84); Y611(0.99) | LDD2238 | [8] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STS-2 Probe Info |
 |
N.A. | LDD0139 | [14] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [15] | |
|
BD-F Probe Info |
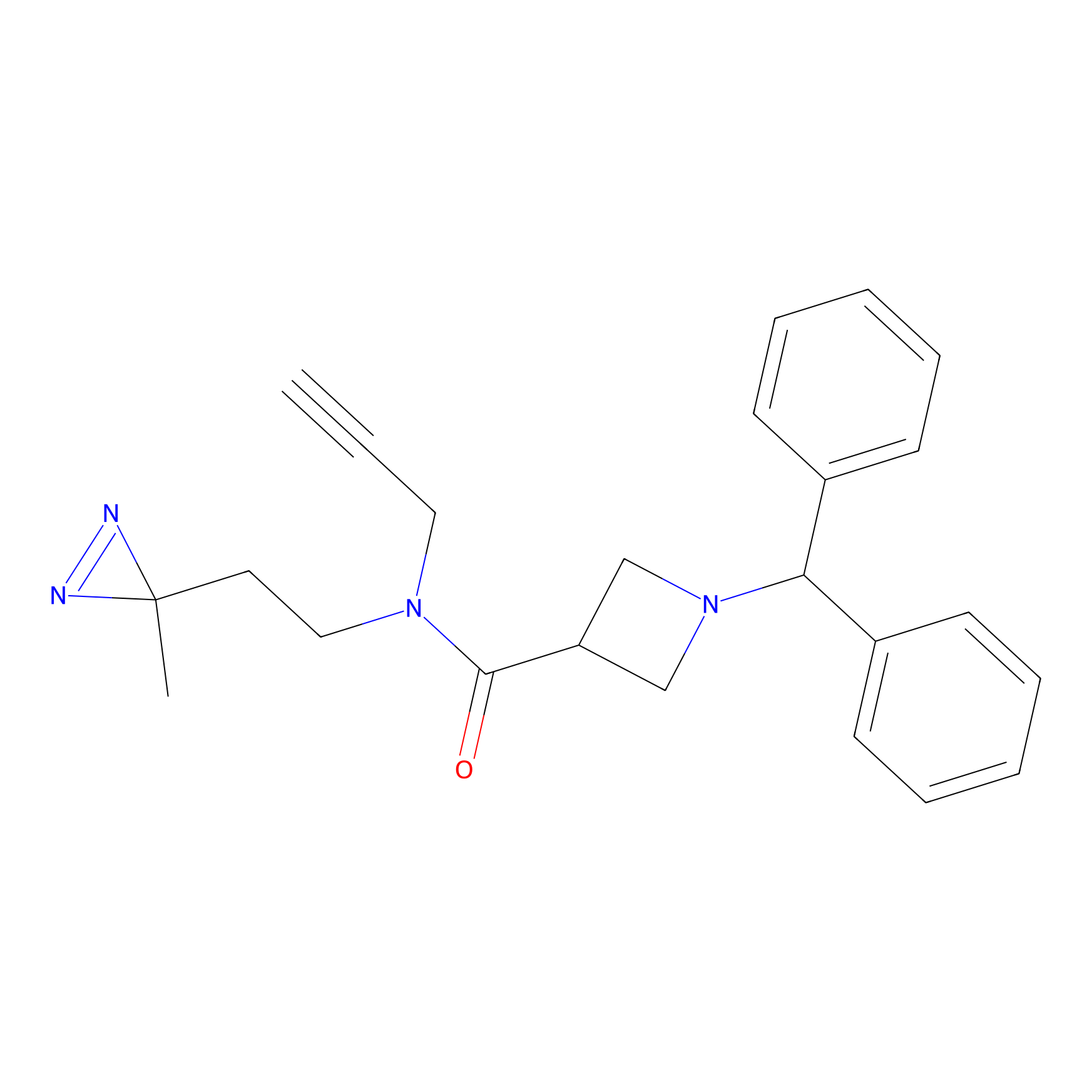 |
G609(0.00); S608(0.00); Y371(0.00); Q612(0.00) | LDD0024 | [16] | |
|
LD-F Probe Info |
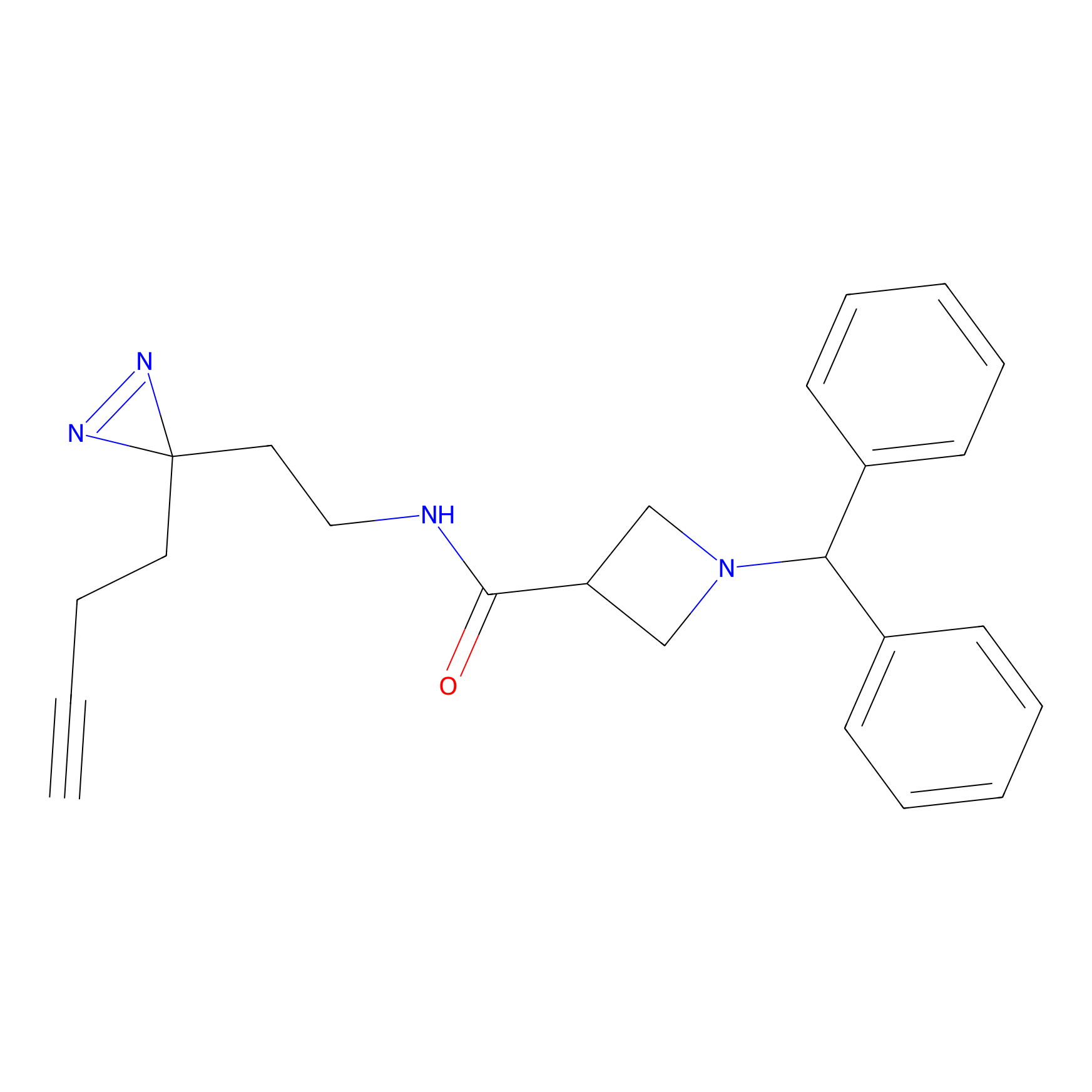 |
G372(0.00); A370(0.00); P365(0.00) | LDD0015 | [16] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0069 | [17] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C306(1.01) | LDD2112 | [5] |
| LDCM0025 | 4SU-RNA | HEK-293T | C603(2.17); C17(2.03) | LDD0168 | [6] |
| LDCM0123 | JWB131 | DM93 | Y107(0.97); Y115(1.18); Y15(1.37); Y183(0.74) | LDD0285 | [7] |
| LDCM0124 | JWB142 | DM93 | Y107(0.93); Y115(0.74); Y15(1.71); Y183(1.76) | LDD0286 | [7] |
| LDCM0125 | JWB146 | DM93 | Y107(0.90); Y115(0.84); Y15(0.79); Y183(0.61) | LDD0287 | [7] |
| LDCM0126 | JWB150 | DM93 | Y107(3.85); Y115(4.84); Y15(4.63); Y183(2.12) | LDD0288 | [7] |
| LDCM0127 | JWB152 | DM93 | Y107(2.62); Y115(2.56); Y15(2.13); Y183(1.38) | LDD0289 | [7] |
| LDCM0128 | JWB198 | DM93 | Y107(1.68); Y115(1.75); Y183(0.11); Y371(0.89) | LDD0290 | [7] |
| LDCM0129 | JWB202 | DM93 | Y107(0.86); Y115(0.85); Y15(0.95); Y183(0.93) | LDD0291 | [7] |
| LDCM0130 | JWB211 | DM93 | Y107(1.19); Y115(0.74); Y15(0.78); Y183(0.38) | LDD0292 | [7] |
| LDCM0023 | KB03 | MDA-MB-231 | C17(0.52) | LDD1701 | [5] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C17(0.84) | LDD2206 | [18] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C17(1.31); C306(0.62) | LDD2207 | [18] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Huntingtin (HTT) | Huntingtin family | P42858 | |||
| Protein S100-B (S100B) | S-100 family | P04271 | |||
Other
References
