Details of the Target
General Information of Target
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TG42 Probe Info |
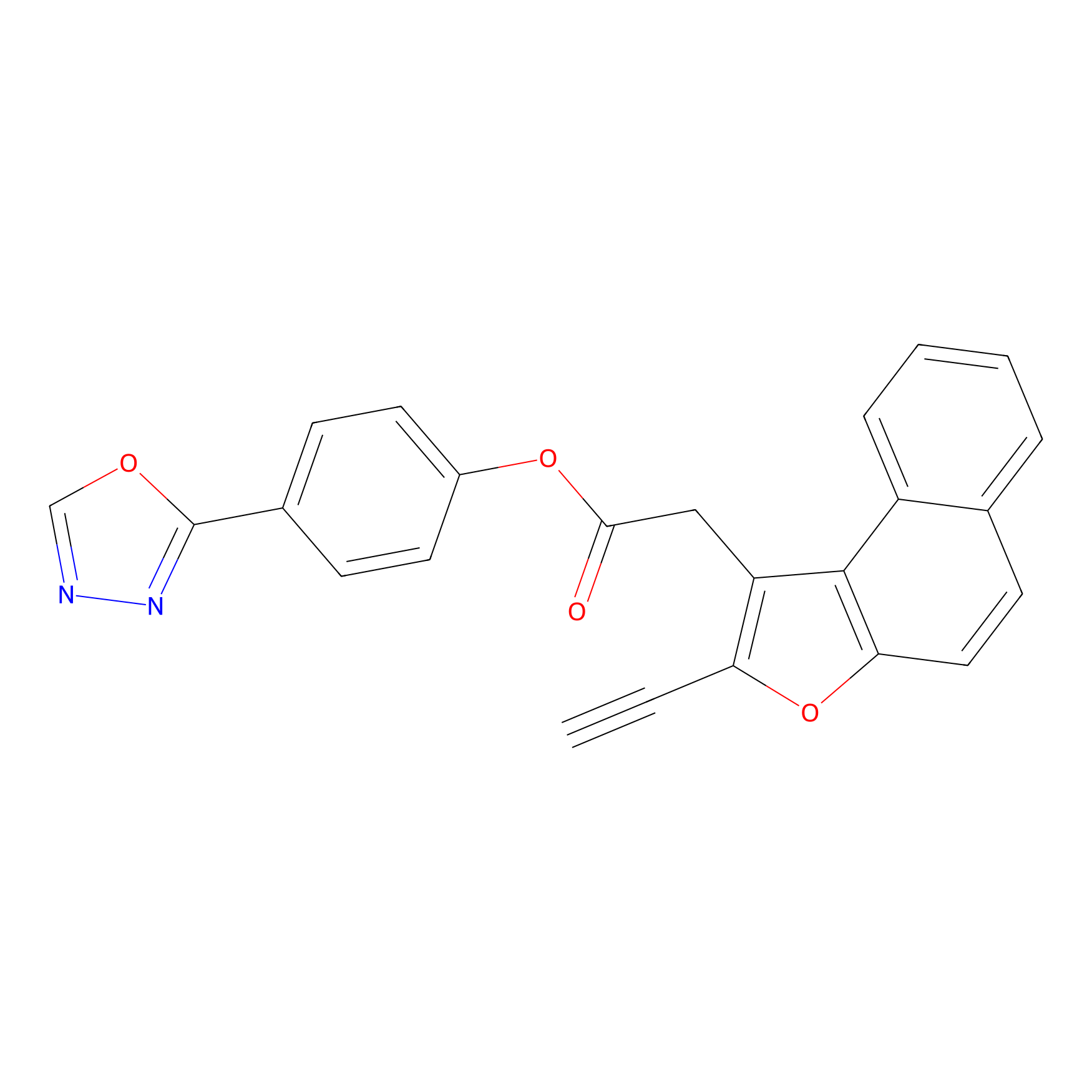 |
4.41 | LDD0043 | [1] | |
|
TH211 Probe Info |
 |
Y90(13.70) | LDD0260 | [2] | |
|
Probe 1 Probe Info |
 |
Y72(97.01); Y161(37.21) | LDD3495 | [3] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0224 | [4] | |
|
AOyne Probe Info |
 |
14.70 | LDD0443 | [5] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C040 Probe Info |
 |
9.06 | LDD1740 | [6] | |
|
C056 Probe Info |
 |
16.91 | LDD1753 | [6] | |
|
C112 Probe Info |
 |
19.97 | LDD1799 | [6] | |
|
C355 Probe Info |
 |
39.67 | LDD2016 | [6] | |
|
C356 Probe Info |
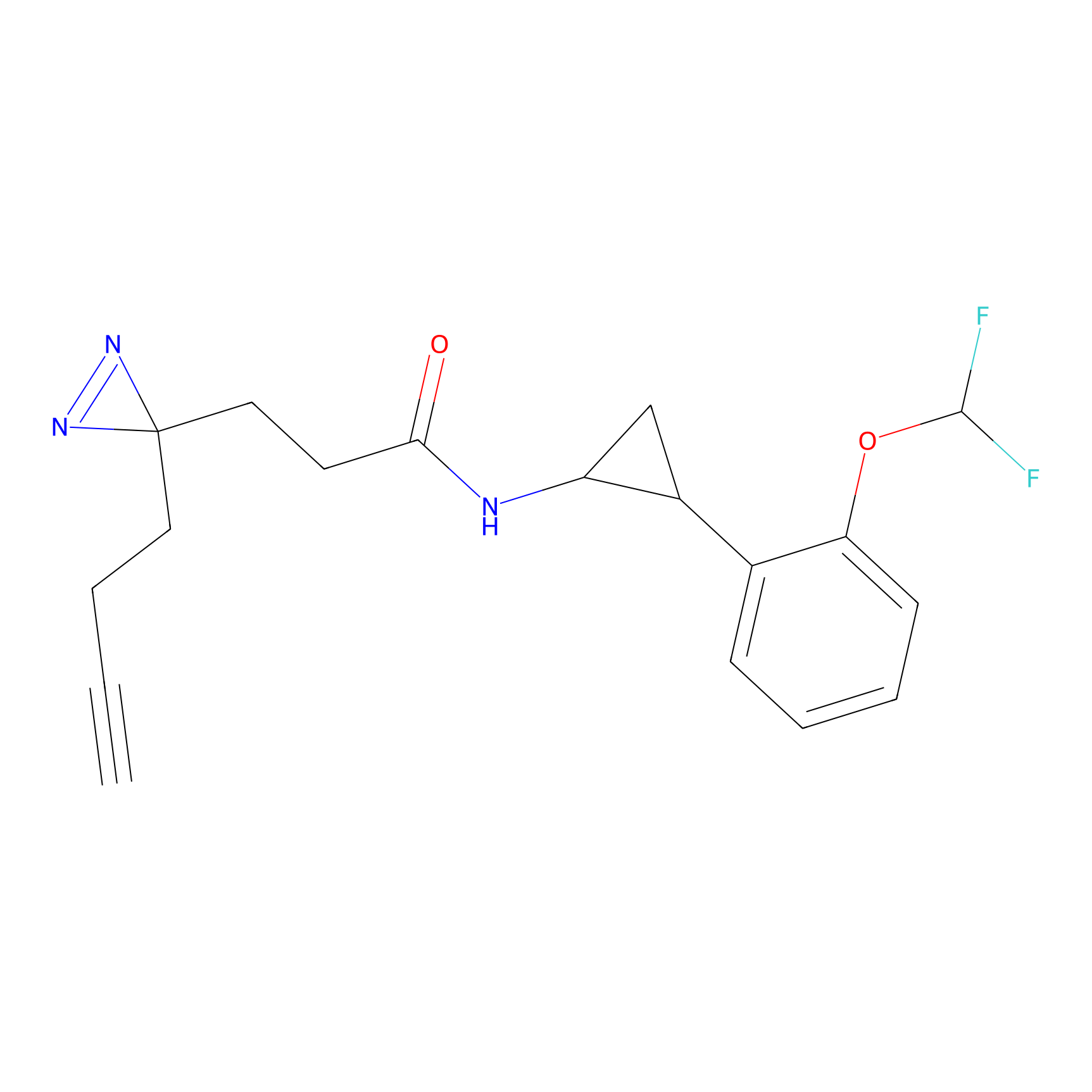 |
15.24 | LDD2017 | [6] | |
|
C357 Probe Info |
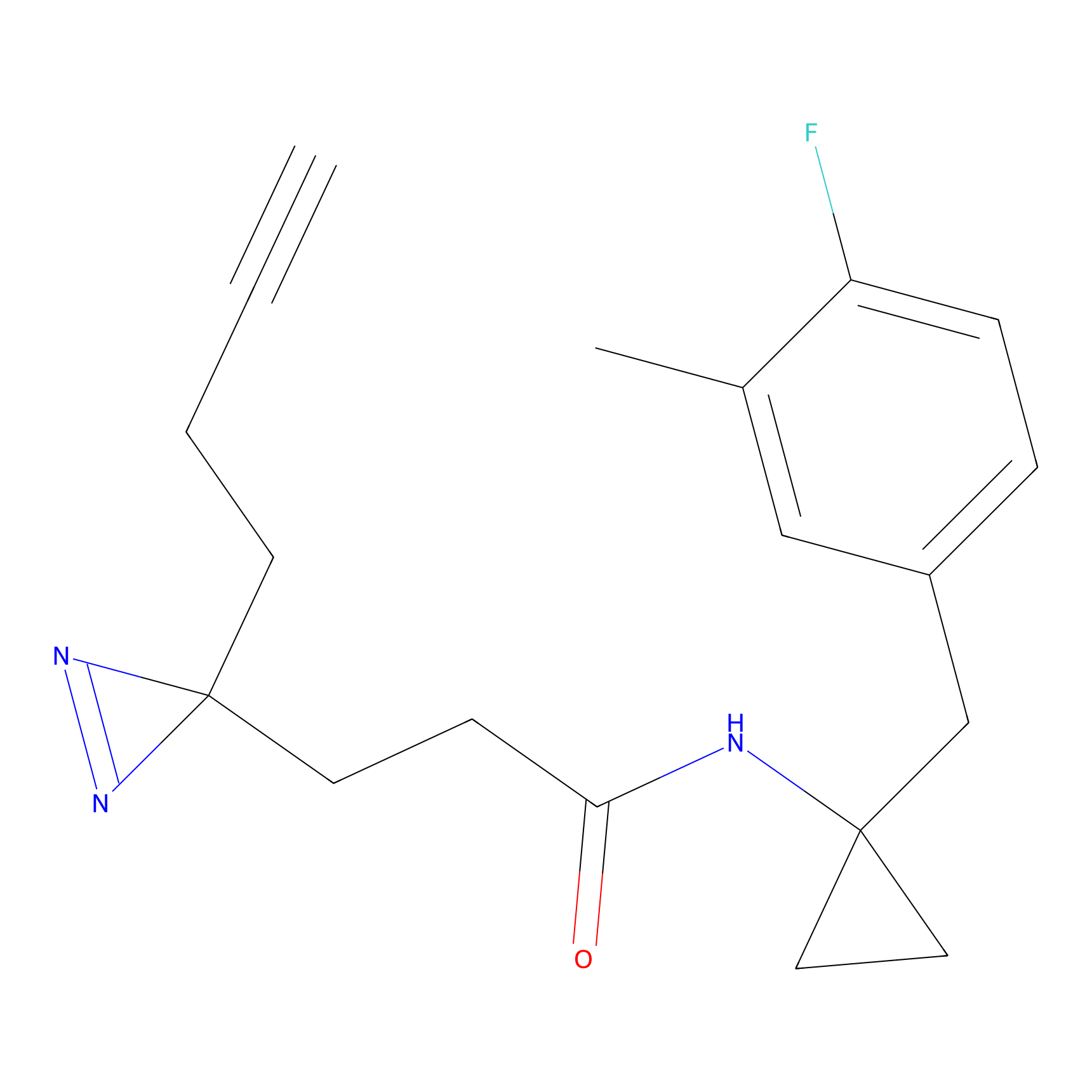 |
7.57 | LDD2018 | [6] | |
|
C360 Probe Info |
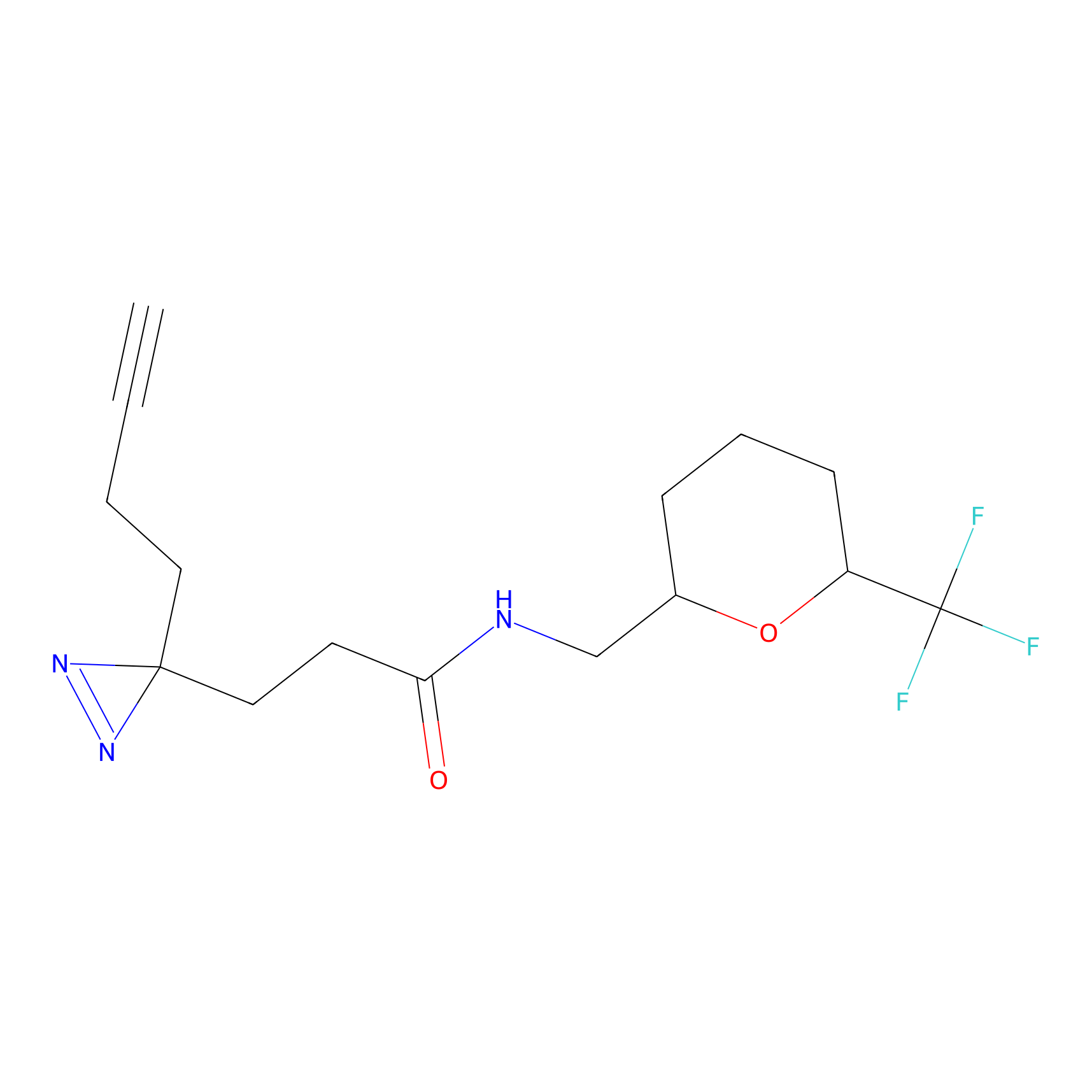 |
5.74 | LDD2021 | [6] | |
|
C361 Probe Info |
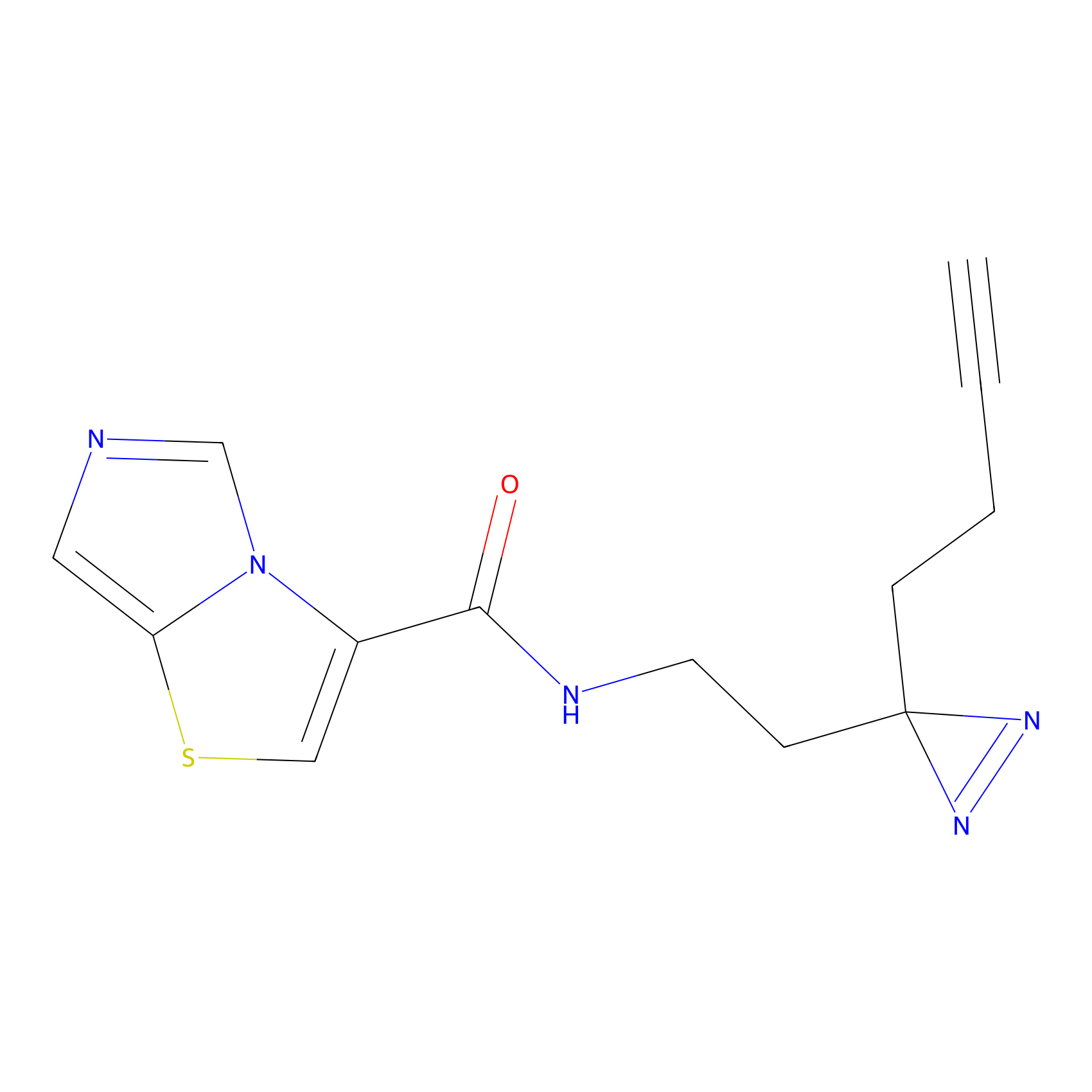 |
20.68 | LDD2022 | [6] | |
|
C390 Probe Info |
 |
25.46 | LDD2049 | [6] | |
|
C413 Probe Info |
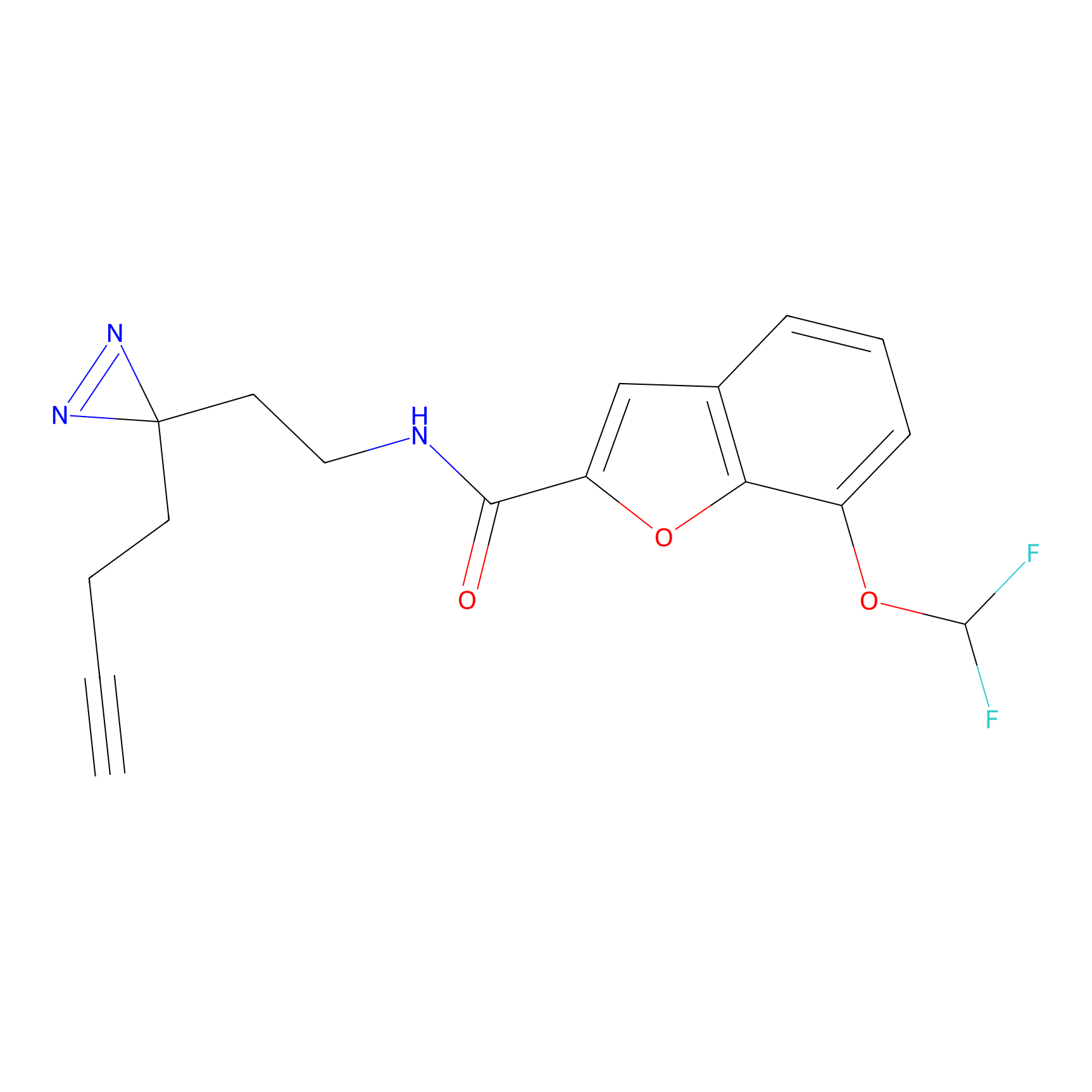 |
12.47 | LDD2069 | [6] | |
|
FFF probe11 Probe Info |
 |
20.00 | LDD0471 | [7] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [7] | |
|
FFF probe14 Probe Info |
 |
20.00 | LDD0477 | [7] | |
|
FFF probe2 Probe Info |
 |
20.00 | LDD0463 | [7] | |
|
OEA-DA Probe Info |
 |
3.24 | LDD0046 | [8] | |
Competitor(s) Related to This Target
The Interaction Atlas With This Target
References
