Details of the Target
General Information of Target
| Target ID | LDTP13532 | |||||
|---|---|---|---|---|---|---|
| Target Name | Malignant T-cell-amplified sequence 1 (MCTS1) | |||||
| Gene Name | MCTS1 | |||||
| Gene ID | 28985 | |||||
| Synonyms |
MCT1; Malignant T-cell-amplified sequence 1; MCT-1; Multiple copies T-cell malignancies |
|||||
| 3D Structure | ||||||
| Sequence |
MLEEDMEVAIKMVVVGNGAVGKSSMIQRYCKGIFTKDYKKTIGVDFLERQIQVNDEDVRL
MLWDTAGQEEFDAITKAYYRGAQACVLVFSTTDRESFEAVSSWREKVVAEVGDIPTVLVQ NKIDLLDDSCIKNEEAEALAKRLKLRFYRTSVKEDLNVNEVFKYLAEKYLQKLKQQIAED PELTHSSSNKIGVFNTSGGSHSGQNSGTLNGGDVINLRPNKQRTKKNRNPFSSCSIP |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
MCTS1 family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Anti-oncogene that plays a role in cell cycle regulation; decreases cell doubling time and anchorage-dependent growth; shortens the duration of G1 transit time and G1/S transition. When constitutively expressed, increases CDK4 and CDK6 kinases activity and CCND1/cyclin D1 protein level, as well as G1 cyclin/CDK complex formation. Involved in translation initiation; promotes recruitment of aminoacetyled initiator tRNA to P site of 40S ribosomes. Can promote release of deacylated tRNA and mRNA from recycled 40S subunits following ABCE1-mediated dissociation of post-termination ribosomal complexes into subunits. Plays a role as translation enhancer; recruits the density-regulated protein/DENR and binds to the cap complex of the 5'-terminus of mRNAs, subsequently altering the mRNA translation profile; up-regulates protein levels of BCL2L2, TFDP1, MRE11, CCND1 and E2F1, while mRNA levels remains constant. Hyperactivates DNA damage signaling pathway; increased gamma-irradiation-induced phosphorylation of histone H2AX, and induces damage foci formation. Increases the overall number of chromosomal abnormalities such as larger chromosomes formation and multiple chromosomal fusions when overexpressed in gamma-irradiated cells. May play a role in promoting lymphoid tumor development: lymphoid cell lines overexpressing MCTS1 exhibit increased growth rates and display increased protection against apoptosis. May contribute to the pathogenesis and progression of breast cancer via promotion of angiogenesis through the decline of inhibitory THBS1/thrombospondin-1, and inhibition of apoptosis. Involved in the process of proteasome degradation to down-regulate Tumor suppressor p53/TP53 in breast cancer cell; Positively regulates phosphorylation of MAPK1 and MAPK3. Involved in translation initiation; promotes aminoacetyled initiator tRNA to P site of 40S ribosomes. Can promote release of deacylated tRNA and mRNA from recycled 40S subunits following ABCE1-mediated dissociation of post-termination ribosomal complexes into subunits.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| HUH28 | SNV: p.H86Y | DBIA Probe Info | |||
| NCIH1155 | SNV: p.E7K | DBIA Probe Info | |||
| RL952 | SNV: p.I111V | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TH211 Probe Info |
 |
Y124(20.00); Y88(20.00); Y169(6.82) | LDD0260 | [1] | |
|
STPyne Probe Info |
 |
K23(10.00); K51(5.28) | LDD0277 | [2] | |
|
AZ-9 Probe Info |
 |
E75(1.58) | LDD2208 | [3] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [2] | |
|
Probe 1 Probe Info |
 |
Y79(14.15) | LDD3495 | [4] | |
|
DBIA Probe Info |
 |
C114(1.23) | LDD3310 | [5] | |
|
BTD Probe Info |
 |
C14(1.43); C144(1.02) | LDD1700 | [6] | |
|
Jackson_14 Probe Info |
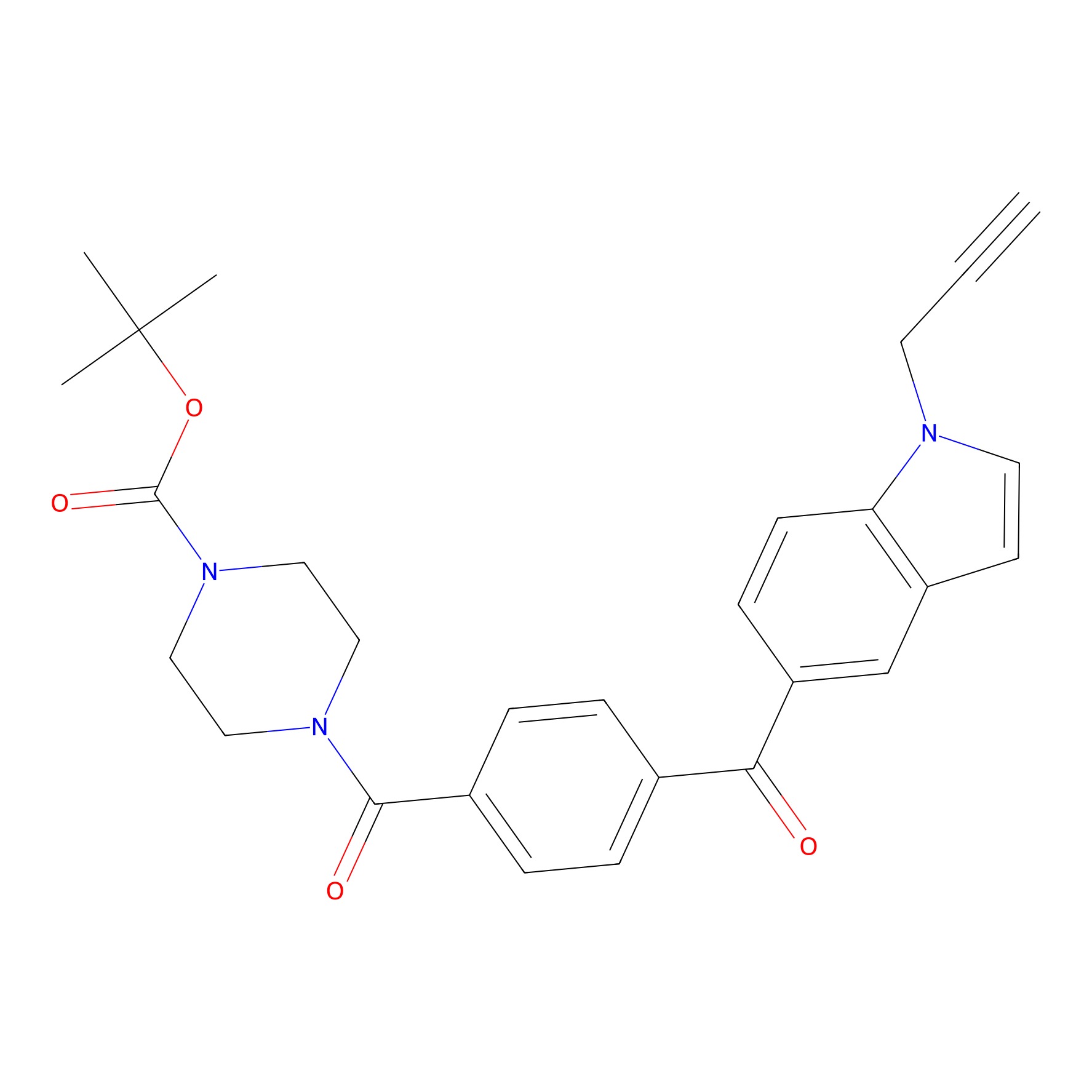 |
2.56 | LDD0123 | [7] | |
|
AHL-Pu-1 Probe Info |
 |
C144(7.94) | LDD0168 | [8] | |
|
HHS-465 Probe Info |
 |
Y169(10.00) | LDD2237 | [9] | |
|
5E-2FA Probe Info |
 |
H58(0.00); H176(0.00); H94(0.00) | LDD2235 | [10] | |
|
ATP probe Probe Info |
 |
K23(0.00); K4(0.00); K8(0.00); K99(0.00) | LDD0199 | [11] | |
|
m-APA Probe Info |
 |
H58(0.00); H94(0.00); H176(0.00) | LDD2231 | [10] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C55(0.00); C14(0.00); C113(0.00); C144(0.00) | LDD0038 | [12] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [13] | |
|
Lodoacetamide azide Probe Info |
 |
C55(0.00); C14(0.00); C113(0.00); C144(0.00) | LDD0037 | [12] | |
|
JW-RF-010 Probe Info |
 |
C14(0.00); C113(0.00) | LDD0026 | [14] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [14] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [15] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [15] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [16] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [16] | |
|
ENE Probe Info |
 |
N.A. | LDD0006 | [15] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [15] | |
|
PF-06672131 Probe Info |
 |
N.A. | LDD0152 | [17] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [15] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [18] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [19] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0357 | [19] | |
|
Acrolein Probe Info |
 |
C113(0.00); C14(0.00); K8(0.00); C144(0.00) | LDD0217 | [20] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [20] | |
|
W1 Probe Info |
 |
C113(0.00); C14(0.00); C144(0.00) | LDD0236 | [21] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [22] | |
|
NAIA_5 Probe Info |
 |
C55(0.00); C14(0.00); C113(0.00); C144(0.00) | LDD2223 | [23] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe14 Probe Info |
 |
11.79 | LDD0477 | [24] | |
|
FFF probe3 Probe Info |
 |
20.00 | LDD0464 | [24] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0502 | 1-(Cyanoacetyl)piperidine | MDA-MB-231 | C14(0.73) | LDD2095 | [6] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C144(1.11); C113(0.94) | LDD2117 | [6] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C14(1.29) | LDD2152 | [6] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C144(0.47) | LDD2132 | [6] |
| LDCM0538 | 4-(Cyanoacetyl)morpholine | MDA-MB-231 | C144(0.74) | LDD2131 | [6] |
| LDCM0025 | 4SU-RNA | HEK-293T | C144(7.94) | LDD0168 | [8] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C144(5.79); C14(2.12) | LDD0169 | [8] |
| LDCM0214 | AC1 | HEK-293T | C144(1.02); C14(0.92); C113(0.92); C55(1.19) | LDD1507 | [25] |
| LDCM0215 | AC10 | HEK-293T | C144(0.98); C14(0.99); C113(0.94) | LDD1508 | [25] |
| LDCM0226 | AC11 | HEK-293T | C144(0.94); C14(0.94); C113(0.92) | LDD1509 | [25] |
| LDCM0237 | AC12 | HEK-293T | C144(0.95); C14(0.90); C113(0.83) | LDD1510 | [25] |
| LDCM0259 | AC14 | HEK-293T | C14(0.93); C113(0.98) | LDD1512 | [25] |
| LDCM0270 | AC15 | HEK-293T | C14(0.92); C113(0.84) | LDD1513 | [25] |
| LDCM0276 | AC17 | HEK-293T | C144(0.96); C14(0.90); C113(0.92); C55(0.97) | LDD1515 | [25] |
| LDCM0277 | AC18 | HEK-293T | C144(0.97); C14(0.98); C113(1.02) | LDD1516 | [25] |
| LDCM0278 | AC19 | HEK-293T | C144(0.79); C14(0.78); C113(0.81) | LDD1517 | [25] |
| LDCM0279 | AC2 | HEK-293T | C144(1.11); C14(1.08); C113(0.94) | LDD1518 | [25] |
| LDCM0280 | AC20 | HEK-293T | C144(0.98); C14(0.94); C113(0.89) | LDD1519 | [25] |
| LDCM0281 | AC21 | HEK-293T | C144(1.05); C14(0.94); C113(0.94) | LDD1520 | [25] |
| LDCM0282 | AC22 | HEK-293T | C14(0.93); C113(1.03) | LDD1521 | [25] |
| LDCM0283 | AC23 | HEK-293T | C14(1.02); C113(0.85) | LDD1522 | [25] |
| LDCM0284 | AC24 | HEK-293T | C144(0.91); C14(1.08); C113(0.98) | LDD1523 | [25] |
| LDCM0285 | AC25 | HEK-293T | C144(1.01); C14(0.91); C113(0.97); C55(0.82) | LDD1524 | [25] |
| LDCM0286 | AC26 | HEK-293T | C144(1.05); C14(0.95); C113(0.92) | LDD1525 | [25] |
| LDCM0287 | AC27 | HEK-293T | C144(0.91); C14(0.95); C113(0.95) | LDD1526 | [25] |
| LDCM0288 | AC28 | HEK-293T | C144(0.96); C14(0.93); C113(0.96) | LDD1527 | [25] |
| LDCM0289 | AC29 | HEK-293T | C144(1.09); C14(0.94); C113(1.06) | LDD1528 | [25] |
| LDCM0290 | AC3 | HEK-293T | C144(0.97); C14(0.91); C113(0.95) | LDD1529 | [25] |
| LDCM0291 | AC30 | HEK-293T | C14(0.95); C113(1.03) | LDD1530 | [25] |
| LDCM0292 | AC31 | HEK-293T | C14(1.00); C113(0.88) | LDD1531 | [25] |
| LDCM0293 | AC32 | HEK-293T | C144(0.90); C14(1.09); C113(0.98) | LDD1532 | [25] |
| LDCM0294 | AC33 | HEK-293T | C144(0.93); C14(0.91); C113(0.90); C55(0.93) | LDD1533 | [25] |
| LDCM0295 | AC34 | HEK-293T | C144(1.02); C14(0.99); C113(0.94) | LDD1534 | [25] |
| LDCM0296 | AC35 | HEK-293T | C144(0.97); C14(0.86); C113(0.89) | LDD1535 | [25] |
| LDCM0297 | AC36 | HEK-293T | C144(0.94); C14(0.85); C113(0.85) | LDD1536 | [25] |
| LDCM0298 | AC37 | HEK-293T | C144(0.93); C14(0.98); C113(1.01) | LDD1537 | [25] |
| LDCM0299 | AC38 | HEK-293T | C14(0.87); C113(1.11) | LDD1538 | [25] |
| LDCM0300 | AC39 | HEK-293T | C14(0.84); C113(0.90) | LDD1539 | [25] |
| LDCM0301 | AC4 | HEK-293T | C144(1.04); C14(0.90); C113(0.90) | LDD1540 | [25] |
| LDCM0302 | AC40 | HEK-293T | C144(0.89); C14(1.19); C113(1.03) | LDD1541 | [25] |
| LDCM0303 | AC41 | HEK-293T | C144(0.92); C14(0.92); C113(0.91); C55(1.08) | LDD1542 | [25] |
| LDCM0304 | AC42 | HEK-293T | C144(0.95); C14(0.97); C113(0.94) | LDD1543 | [25] |
| LDCM0305 | AC43 | HEK-293T | C144(0.94); C14(0.86); C113(0.95) | LDD1544 | [25] |
| LDCM0306 | AC44 | HEK-293T | C144(0.95); C14(0.95); C113(0.92) | LDD1545 | [25] |
| LDCM0307 | AC45 | HEK-293T | C144(0.99); C14(0.89); C113(0.98) | LDD1546 | [25] |
| LDCM0308 | AC46 | HEK-293T | C14(0.87); C113(1.06) | LDD1547 | [25] |
| LDCM0309 | AC47 | HEK-293T | C14(0.85); C113(0.87) | LDD1548 | [25] |
| LDCM0310 | AC48 | HEK-293T | C144(0.91); C14(1.21); C113(0.98) | LDD1549 | [25] |
| LDCM0311 | AC49 | HEK-293T | C144(0.92); C14(0.93); C113(0.81); C55(0.83) | LDD1550 | [25] |
| LDCM0312 | AC5 | HEK-293T | C144(1.22); C14(0.98); C113(1.04) | LDD1551 | [25] |
| LDCM0313 | AC50 | HEK-293T | C144(1.02); C14(1.02); C113(1.01) | LDD1552 | [25] |
| LDCM0314 | AC51 | HEK-293T | C144(1.01); C14(0.92); C113(1.00) | LDD1553 | [25] |
| LDCM0315 | AC52 | HEK-293T | C144(0.96); C14(0.92); C113(0.84) | LDD1554 | [25] |
| LDCM0316 | AC53 | HEK-293T | C144(1.03); C14(0.94); C113(0.95) | LDD1555 | [25] |
| LDCM0317 | AC54 | HEK-293T | C14(0.86); C113(1.10) | LDD1556 | [25] |
| LDCM0318 | AC55 | HEK-293T | C14(0.88); C113(0.81) | LDD1557 | [25] |
| LDCM0319 | AC56 | HEK-293T | C144(0.86); C14(1.14); C113(0.96) | LDD1558 | [25] |
| LDCM0320 | AC57 | HEK-293T | C144(0.96); C14(0.94); C113(0.86); C55(1.23) | LDD1559 | [25] |
| LDCM0321 | AC58 | HEK-293T | C144(1.04); C14(1.03); C113(0.95) | LDD1560 | [25] |
| LDCM0322 | AC59 | HEK-293T | C144(0.96); C14(0.92); C113(0.96) | LDD1561 | [25] |
| LDCM0323 | AC6 | HEK-293T | C14(0.89); C113(1.11) | LDD1562 | [25] |
| LDCM0324 | AC60 | HEK-293T | C144(0.98); C14(0.91); C113(0.82) | LDD1563 | [25] |
| LDCM0325 | AC61 | HEK-293T | C144(1.01); C14(0.91); C113(1.01) | LDD1564 | [25] |
| LDCM0326 | AC62 | HEK-293T | C14(0.92); C113(1.07) | LDD1565 | [25] |
| LDCM0327 | AC63 | HEK-293T | C14(0.86); C113(0.88) | LDD1566 | [25] |
| LDCM0328 | AC64 | HEK-293T | C144(1.02); C14(1.10); C113(0.99) | LDD1567 | [25] |
| LDCM0334 | AC7 | HEK-293T | C14(0.99); C113(0.88) | LDD1568 | [25] |
| LDCM0345 | AC8 | HEK-293T | C144(0.96); C14(1.21); C113(0.99) | LDD1569 | [25] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C14(1.08) | LDD2113 | [6] |
| LDCM0248 | AKOS034007472 | HEK-293T | C144(1.04); C14(0.99); C113(0.95) | LDD1511 | [25] |
| LDCM0356 | AKOS034007680 | HEK-293T | C144(0.97); C14(0.93); C113(0.88); C55(1.00) | LDD1570 | [25] |
| LDCM0275 | AKOS034007705 | HEK-293T | C144(0.92); C14(1.07); C113(0.99) | LDD1514 | [25] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C144(0.78); C113(0.87) | LDD2091 | [6] |
| LDCM0108 | Chloroacetamide | HeLa | C113(0.00); C14(0.00) | LDD0222 | [20] |
| LDCM0632 | CL-Sc | Hep-G2 | C113(3.07); C14(1.10); C14(0.98) | LDD2227 | [23] |
| LDCM0367 | CL1 | HEK-293T | C144(1.14); C14(0.90); C113(1.07) | LDD1571 | [25] |
| LDCM0368 | CL10 | HEK-293T | C14(0.75); C113(1.01) | LDD1572 | [25] |
| LDCM0369 | CL100 | HEK-293T | C144(0.99); C14(0.91); C113(0.93) | LDD1573 | [25] |
| LDCM0370 | CL101 | HEK-293T | C144(1.01); C14(0.95); C113(0.98) | LDD1574 | [25] |
| LDCM0371 | CL102 | HEK-293T | C144(1.01); C14(0.85); C113(0.92) | LDD1575 | [25] |
| LDCM0372 | CL103 | HEK-293T | C144(0.99); C14(0.95); C113(0.95) | LDD1576 | [25] |
| LDCM0373 | CL104 | HEK-293T | C144(1.04); C14(0.90); C113(0.92) | LDD1577 | [25] |
| LDCM0374 | CL105 | HEK-293T | C144(1.05); C14(0.95); C113(0.85) | LDD1578 | [25] |
| LDCM0375 | CL106 | HEK-293T | C144(1.06); C14(0.85); C113(0.93) | LDD1579 | [25] |
| LDCM0376 | CL107 | HEK-293T | C144(0.97); C14(1.01); C113(0.99) | LDD1580 | [25] |
| LDCM0377 | CL108 | HEK-293T | C144(1.01); C14(0.92); C113(0.91) | LDD1581 | [25] |
| LDCM0378 | CL109 | HEK-293T | C144(1.03); C14(0.92); C113(0.99) | LDD1582 | [25] |
| LDCM0379 | CL11 | HEK-293T | C14(0.93); C113(1.00) | LDD1583 | [25] |
| LDCM0380 | CL110 | HEK-293T | C144(1.01); C14(0.81); C113(0.90) | LDD1584 | [25] |
| LDCM0381 | CL111 | HEK-293T | C144(0.91); C14(0.99); C113(0.93) | LDD1585 | [25] |
| LDCM0382 | CL112 | HEK-293T | C144(1.01); C14(0.84); C113(0.90) | LDD1586 | [25] |
| LDCM0383 | CL113 | HEK-293T | C144(0.99); C14(0.97); C113(1.00) | LDD1587 | [25] |
| LDCM0384 | CL114 | HEK-293T | C144(1.05); C14(0.76); C113(0.90) | LDD1588 | [25] |
| LDCM0385 | CL115 | HEK-293T | C144(0.95); C14(0.94); C113(0.96) | LDD1589 | [25] |
| LDCM0386 | CL116 | HEK-293T | C144(0.95); C14(0.82); C113(0.95) | LDD1590 | [25] |
| LDCM0387 | CL117 | HEK-293T | C144(1.00); C14(0.92); C113(0.93) | LDD1591 | [25] |
| LDCM0388 | CL118 | HEK-293T | C144(1.00); C14(0.83); C113(0.89) | LDD1592 | [25] |
| LDCM0389 | CL119 | HEK-293T | C144(0.98); C14(0.96); C113(1.03) | LDD1593 | [25] |
| LDCM0390 | CL12 | HEK-293T | C144(0.82); C14(1.07); C113(0.91) | LDD1594 | [25] |
| LDCM0391 | CL120 | HEK-293T | C144(0.96); C14(0.84); C113(0.88) | LDD1595 | [25] |
| LDCM0392 | CL121 | HEK-293T | C144(1.04); C14(0.95); C113(0.91) | LDD1596 | [25] |
| LDCM0393 | CL122 | HEK-293T | C144(1.03); C14(0.89); C113(0.92) | LDD1597 | [25] |
| LDCM0394 | CL123 | HEK-293T | C144(0.89); C14(0.91); C113(0.98) | LDD1598 | [25] |
| LDCM0395 | CL124 | HEK-293T | C144(0.97); C14(0.85); C113(0.86) | LDD1599 | [25] |
| LDCM0396 | CL125 | HEK-293T | C144(1.15); C14(0.94); C113(1.00) | LDD1600 | [25] |
| LDCM0397 | CL126 | HEK-293T | C144(0.96); C14(0.84); C113(0.93) | LDD1601 | [25] |
| LDCM0398 | CL127 | HEK-293T | C144(0.97); C14(0.98); C113(0.93) | LDD1602 | [25] |
| LDCM0399 | CL128 | HEK-293T | C144(0.99); C14(0.86); C113(0.96) | LDD1603 | [25] |
| LDCM0400 | CL13 | HEK-293T | C144(1.13); C14(0.92); C113(0.98) | LDD1604 | [25] |
| LDCM0401 | CL14 | HEK-293T | C144(1.05); C14(0.97); C113(0.98) | LDD1605 | [25] |
| LDCM0402 | CL15 | HEK-293T | C144(0.89); C14(0.94); C113(0.94) | LDD1606 | [25] |
| LDCM0403 | CL16 | HEK-293T | C144(1.04); C14(1.02); C113(0.95) | LDD1607 | [25] |
| LDCM0404 | CL17 | HEK-293T | C144(0.93); C14(0.82); C113(0.93); C55(0.86) | LDD1608 | [25] |
| LDCM0405 | CL18 | HEK-293T | C144(0.95); C14(1.06); C113(0.97) | LDD1609 | [25] |
| LDCM0406 | CL19 | HEK-293T | C144(1.00); C14(0.94); C113(0.94) | LDD1610 | [25] |
| LDCM0407 | CL2 | HEK-293T | C144(1.15); C14(1.01); C113(0.99) | LDD1611 | [25] |
| LDCM0408 | CL20 | HEK-293T | C144(0.91); C14(0.91); C113(0.84) | LDD1612 | [25] |
| LDCM0409 | CL21 | HEK-293T | C144(0.87); C14(0.89); C113(0.91) | LDD1613 | [25] |
| LDCM0410 | CL22 | HEK-293T | C14(0.99); C113(1.10) | LDD1614 | [25] |
| LDCM0411 | CL23 | HEK-293T | C14(1.02); C113(1.00) | LDD1615 | [25] |
| LDCM0412 | CL24 | HEK-293T | C144(0.78); C14(1.16); C113(1.05) | LDD1616 | [25] |
| LDCM0413 | CL25 | HEK-293T | C144(1.06); C14(0.90); C113(1.05) | LDD1617 | [25] |
| LDCM0414 | CL26 | HEK-293T | C144(1.06); C14(0.93); C113(0.99) | LDD1618 | [25] |
| LDCM0415 | CL27 | HEK-293T | C144(0.97); C14(0.99); C113(0.96) | LDD1619 | [25] |
| LDCM0416 | CL28 | HEK-293T | C144(1.00); C14(0.97); C113(0.88) | LDD1620 | [25] |
| LDCM0417 | CL29 | HEK-293T | C144(1.03); C14(0.96); C113(0.92); C55(1.00) | LDD1621 | [25] |
| LDCM0418 | CL3 | HEK-293T | C144(1.00); C14(1.04); C113(0.95) | LDD1622 | [25] |
| LDCM0419 | CL30 | HEK-293T | C144(1.00); C14(1.07); C113(1.00) | LDD1623 | [25] |
| LDCM0420 | CL31 | HEK-293T | C144(0.91); C14(0.92); C113(0.90) | LDD1624 | [25] |
| LDCM0421 | CL32 | HEK-293T | C144(0.92); C14(0.97); C113(0.87) | LDD1625 | [25] |
| LDCM0422 | CL33 | HEK-293T | C144(1.00); C14(0.82); C113(1.02) | LDD1626 | [25] |
| LDCM0423 | CL34 | HEK-293T | C14(0.95); C113(1.03) | LDD1627 | [25] |
| LDCM0424 | CL35 | HEK-293T | C14(0.96); C113(0.85) | LDD1628 | [25] |
| LDCM0425 | CL36 | HEK-293T | C144(0.78); C14(1.08); C113(0.95) | LDD1629 | [25] |
| LDCM0426 | CL37 | HEK-293T | C144(1.11); C14(0.98); C113(1.02) | LDD1630 | [25] |
| LDCM0428 | CL39 | HEK-293T | C144(1.03); C14(0.96); C113(1.03) | LDD1632 | [25] |
| LDCM0429 | CL4 | HEK-293T | C144(1.02); C14(0.94); C113(0.91) | LDD1633 | [25] |
| LDCM0430 | CL40 | HEK-293T | C144(1.04); C14(0.94); C113(0.93) | LDD1634 | [25] |
| LDCM0431 | CL41 | HEK-293T | C144(1.02); C14(0.90); C113(0.92); C55(0.85) | LDD1635 | [25] |
| LDCM0432 | CL42 | HEK-293T | C144(1.07); C14(1.01); C113(1.00) | LDD1636 | [25] |
| LDCM0433 | CL43 | HEK-293T | C144(1.01); C14(1.02); C113(0.96) | LDD1637 | [25] |
| LDCM0434 | CL44 | HEK-293T | C144(0.93); C14(0.97); C113(0.92) | LDD1638 | [25] |
| LDCM0435 | CL45 | HEK-293T | C144(1.02); C14(0.91); C113(1.06) | LDD1639 | [25] |
| LDCM0436 | CL46 | HEK-293T | C14(0.96); C113(1.07) | LDD1640 | [25] |
| LDCM0437 | CL47 | HEK-293T | C14(1.01); C113(0.85) | LDD1641 | [25] |
| LDCM0438 | CL48 | HEK-293T | C144(0.86); C14(1.08); C113(0.98) | LDD1642 | [25] |
| LDCM0439 | CL49 | HEK-293T | C144(1.16); C14(0.96); C113(1.05) | LDD1643 | [25] |
| LDCM0440 | CL5 | HEK-293T | C144(0.95); C14(1.03); C113(0.96); C55(1.20) | LDD1644 | [25] |
| LDCM0441 | CL50 | HEK-293T | C144(1.04); C14(0.87); C113(0.98) | LDD1645 | [25] |
| LDCM0443 | CL52 | HEK-293T | C144(0.94); C14(0.87); C113(0.90) | LDD1646 | [25] |
| LDCM0444 | CL53 | HEK-293T | C144(0.89); C14(0.82); C113(0.89); C55(0.87) | LDD1647 | [25] |
| LDCM0445 | CL54 | HEK-293T | C144(0.98); C14(0.87); C113(0.91) | LDD1648 | [25] |
| LDCM0446 | CL55 | HEK-293T | C144(0.96); C14(0.87); C113(0.94) | LDD1649 | [25] |
| LDCM0447 | CL56 | HEK-293T | C144(0.93); C14(0.87); C113(0.83) | LDD1650 | [25] |
| LDCM0448 | CL57 | HEK-293T | C144(0.91); C14(0.77); C113(1.06) | LDD1651 | [25] |
| LDCM0449 | CL58 | HEK-293T | C14(0.84); C113(1.03) | LDD1652 | [25] |
| LDCM0450 | CL59 | HEK-293T | C14(0.91); C113(0.86) | LDD1653 | [25] |
| LDCM0451 | CL6 | HEK-293T | C144(0.96); C14(0.97); C113(1.01) | LDD1654 | [25] |
| LDCM0452 | CL60 | HEK-293T | C144(0.82); C14(1.17); C113(1.01) | LDD1655 | [25] |
| LDCM0453 | CL61 | HEK-293T | C144(1.18); C14(0.99); C113(1.12) | LDD1656 | [25] |
| LDCM0454 | CL62 | HEK-293T | C144(1.10); C14(0.91); C113(0.99) | LDD1657 | [25] |
| LDCM0455 | CL63 | HEK-293T | C144(0.94); C14(1.01); C113(0.96) | LDD1658 | [25] |
| LDCM0456 | CL64 | HEK-293T | C144(0.95); C14(0.85); C113(0.95) | LDD1659 | [25] |
| LDCM0457 | CL65 | HEK-293T | C144(0.99); C14(0.93); C113(0.87); C55(0.98) | LDD1660 | [25] |
| LDCM0458 | CL66 | HEK-293T | C144(0.98); C14(1.02); C113(0.98) | LDD1661 | [25] |
| LDCM0459 | CL67 | HEK-293T | C144(0.94); C14(0.90); C113(0.88) | LDD1662 | [25] |
| LDCM0460 | CL68 | HEK-293T | C144(0.93); C14(0.91); C113(0.89) | LDD1663 | [25] |
| LDCM0461 | CL69 | HEK-293T | C144(0.85); C14(0.91); C113(1.04) | LDD1664 | [25] |
| LDCM0462 | CL7 | HEK-293T | C144(1.05); C14(0.94); C113(1.04) | LDD1665 | [25] |
| LDCM0463 | CL70 | HEK-293T | C14(0.96); C113(1.05) | LDD1666 | [25] |
| LDCM0464 | CL71 | HEK-293T | C14(0.94); C113(0.83) | LDD1667 | [25] |
| LDCM0465 | CL72 | HEK-293T | C144(0.82); C14(1.19); C113(1.00) | LDD1668 | [25] |
| LDCM0466 | CL73 | HEK-293T | C144(1.10); C14(0.94); C113(1.08) | LDD1669 | [25] |
| LDCM0467 | CL74 | HEK-293T | C144(1.04); C14(0.92); C113(0.99) | LDD1670 | [25] |
| LDCM0469 | CL76 | HEK-293T | C144(0.99); C14(0.93); C113(0.90) | LDD1672 | [25] |
| LDCM0470 | CL77 | HEK-293T | C144(0.92); C14(0.85); C113(0.94); C55(1.31) | LDD1673 | [25] |
| LDCM0471 | CL78 | HEK-293T | C144(0.97); C14(1.04); C113(1.02) | LDD1674 | [25] |
| LDCM0472 | CL79 | HEK-293T | C144(1.02); C14(0.98); C113(0.97) | LDD1675 | [25] |
| LDCM0473 | CL8 | HEK-293T | C144(0.91); C14(0.65); C113(0.79) | LDD1676 | [25] |
| LDCM0474 | CL80 | HEK-293T | C144(0.96); C14(0.93); C113(0.94) | LDD1677 | [25] |
| LDCM0475 | CL81 | HEK-293T | C144(0.88); C14(0.98); C113(0.96) | LDD1678 | [25] |
| LDCM0476 | CL82 | HEK-293T | C14(0.92); C113(1.04) | LDD1679 | [25] |
| LDCM0477 | CL83 | HEK-293T | C14(0.90); C113(0.79) | LDD1680 | [25] |
| LDCM0478 | CL84 | HEK-293T | C144(0.91); C14(1.08); C113(0.95) | LDD1681 | [25] |
| LDCM0479 | CL85 | HEK-293T | C144(1.31); C14(1.02); C113(1.08) | LDD1682 | [25] |
| LDCM0480 | CL86 | HEK-293T | C144(1.25); C14(0.88); C113(0.97) | LDD1683 | [25] |
| LDCM0481 | CL87 | HEK-293T | C144(0.99); C14(0.97); C113(0.94) | LDD1684 | [25] |
| LDCM0482 | CL88 | HEK-293T | C144(1.01); C14(0.92); C113(0.93) | LDD1685 | [25] |
| LDCM0483 | CL89 | HEK-293T | C144(0.98); C14(0.97); C113(0.93); C55(1.28) | LDD1686 | [25] |
| LDCM0484 | CL9 | HEK-293T | C144(0.84); C14(0.93); C113(1.09) | LDD1687 | [25] |
| LDCM0485 | CL90 | HEK-293T | C144(0.95); C14(0.79); C113(0.90) | LDD1688 | [25] |
| LDCM0486 | CL91 | HEK-293T | C144(1.08); C14(1.00); C113(0.97) | LDD1689 | [25] |
| LDCM0487 | CL92 | HEK-293T | C144(0.95); C14(0.87); C113(0.84) | LDD1690 | [25] |
| LDCM0488 | CL93 | HEK-293T | C144(1.18); C14(0.86); C113(0.99) | LDD1691 | [25] |
| LDCM0489 | CL94 | HEK-293T | C14(0.86); C113(1.08) | LDD1692 | [25] |
| LDCM0490 | CL95 | HEK-293T | C14(0.76); C113(0.83) | LDD1693 | [25] |
| LDCM0491 | CL96 | HEK-293T | C144(1.26); C14(1.10); C113(0.99) | LDD1694 | [25] |
| LDCM0492 | CL97 | HEK-293T | C144(1.04); C14(0.89); C113(0.95) | LDD1695 | [25] |
| LDCM0493 | CL98 | HEK-293T | C144(1.00); C14(0.88); C113(0.89) | LDD1696 | [25] |
| LDCM0494 | CL99 | HEK-293T | C144(1.03); C14(1.04); C113(0.99) | LDD1697 | [25] |
| LDCM0191 | Compound 21 | HEK-293T | 10.10 | LDD0508 | [24] |
| LDCM0192 | Compound 35 | HEK-293T | 6.35 | LDD0509 | [24] |
| LDCM0193 | Compound 36 | HEK-293T | 8.20 | LDD0511 | [24] |
| LDCM0634 | CY-0357 | Hep-G2 | C14(1.09); C14(0.65) | LDD2228 | [23] |
| LDCM0495 | E2913 | HEK-293T | C144(1.06); C14(0.96); C113(0.97) | LDD1698 | [25] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C113(2.06); C14(1.32) | LDD1702 | [6] |
| LDCM0625 | F8 | Ramos | C14(2.24); C113(0.71) | LDD2187 | [26] |
| LDCM0572 | Fragment10 | Ramos | C14(0.82); C113(0.57) | LDD2189 | [26] |
| LDCM0573 | Fragment11 | Ramos | C14(0.02); C113(0.25) | LDD2190 | [26] |
| LDCM0574 | Fragment12 | Ramos | C14(0.78); C113(0.59) | LDD2191 | [26] |
| LDCM0575 | Fragment13 | Ramos | C14(0.85); C113(1.29) | LDD2192 | [26] |
| LDCM0576 | Fragment14 | Ramos | C14(0.89); C113(0.73) | LDD2193 | [26] |
| LDCM0579 | Fragment20 | Ramos | C14(0.68); C113(0.55) | LDD2194 | [26] |
| LDCM0580 | Fragment21 | Ramos | C14(0.86); C113(0.95) | LDD2195 | [26] |
| LDCM0582 | Fragment23 | Ramos | C14(0.87); C113(1.06) | LDD2196 | [26] |
| LDCM0578 | Fragment27 | Ramos | C14(0.87); C113(1.01) | LDD2197 | [26] |
| LDCM0586 | Fragment28 | Ramos | C14(0.81); C113(1.22) | LDD2198 | [26] |
| LDCM0588 | Fragment30 | Ramos | C14(0.89); C113(1.18) | LDD2199 | [26] |
| LDCM0589 | Fragment31 | Ramos | C14(0.83); C113(1.58) | LDD2200 | [26] |
| LDCM0590 | Fragment32 | Ramos | C14(0.68); C113(0.51) | LDD2201 | [26] |
| LDCM0468 | Fragment33 | HEK-293T | C144(1.00); C14(0.99); C113(0.97) | LDD1671 | [25] |
| LDCM0596 | Fragment38 | Ramos | C14(0.75); C113(0.85) | LDD2203 | [26] |
| LDCM0566 | Fragment4 | Ramos | C14(0.82); C113(0.87) | LDD2184 | [26] |
| LDCM0427 | Fragment51 | HEK-293T | C144(1.07); C14(0.94); C113(0.94) | LDD1631 | [25] |
| LDCM0610 | Fragment52 | Ramos | C14(1.12); C113(1.14) | LDD2204 | [26] |
| LDCM0614 | Fragment56 | Ramos | C14(0.93); C113(1.01) | LDD2205 | [26] |
| LDCM0569 | Fragment7 | Ramos | C14(0.74); C113(0.46) | LDD2186 | [26] |
| LDCM0571 | Fragment9 | Ramos | C14(0.60); C113(0.43) | LDD2188 | [26] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [20] |
| LDCM0022 | KB02 | HEK-293T | C14(1.00); C113(0.99); C144(1.01) | LDD1492 | [25] |
| LDCM0023 | KB03 | HEK-293T | C14(0.99); C113(1.10); C144(0.97) | LDD1497 | [25] |
| LDCM0024 | KB05 | COLO792 | C114(1.23) | LDD3310 | [5] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0225 | [20] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C14(0.20); C113(0.93) | LDD2089 | [6] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C14(2.89) | LDD2090 | [6] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C14(0.92) | LDD2093 | [6] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C144(2.67) | LDD2094 | [6] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C14(1.22) | LDD2097 | [6] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C144(1.01) | LDD2098 | [6] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C144(1.77); C113(0.94) | LDD2099 | [6] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C14(1.31) | LDD2101 | [6] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C113(0.70) | LDD2106 | [6] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C14(0.91); C144(1.04); C113(0.88) | LDD2107 | [6] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C14(0.59); C144(0.70) | LDD2109 | [6] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C14(1.01); C144(1.76) | LDD2111 | [6] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C14(0.94) | LDD2115 | [6] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C144(1.16) | LDD2119 | [6] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C144(0.63) | LDD2120 | [6] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C113(0.92) | LDD2123 | [6] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C14(0.93); C144(1.01) | LDD2125 | [6] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C14(0.94) | LDD2127 | [6] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C113(0.83) | LDD2128 | [6] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C14(0.76) | LDD2129 | [6] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C14(0.78); C144(0.62) | LDD2133 | [6] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C14(0.77) | LDD2134 | [6] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C14(0.97); C144(0.65) | LDD2135 | [6] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C14(1.02); C144(1.17); C113(0.81) | LDD2136 | [6] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C144(0.98); C113(0.94) | LDD2137 | [6] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C14(1.43); C144(1.02) | LDD1700 | [6] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C14(1.02); C144(1.19) | LDD2140 | [6] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C14(3.00) | LDD2143 | [6] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C144(1.18) | LDD2144 | [6] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C14(0.84); C144(0.80) | LDD2146 | [6] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C14(0.85) | LDD2148 | [6] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C14(0.97) | LDD2206 | [27] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C14(0.68); C14(0.11) | LDD2207 | [27] |
| LDCM0131 | RA190 | MM1.R | C14(1.30) | LDD0304 | [28] |
| LDCM0016 | Ranjitkar_cp1 | MDA-MB-231 | 2.56 | LDD0123 | [7] |
The Interaction Atlas With This Target
References
