Details of the Target
General Information of Target
| Target ID | LDTP16201 | |||||
|---|---|---|---|---|---|---|
| Target Name | B-cell receptor-associated protein 29 (BCAP29) | |||||
| Gene Name | BCAP29 | |||||
| Gene ID | 55973 | |||||
| Synonyms |
BAP29; B-cell receptor-associated protein 29; BCR-associated protein 29; Bap29 |
|||||
| 3D Structure | ||||||
| Sequence |
MNMSKQPVSNVRAIQANINIPMGAFRPGAGQPPRRKECTPEVEEGVPPTSDEEKKPIPGA
KKLPGPAVNLSEIQNIKSELKYVPKAEQ |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
BCAP29/BCAP31 family
|
|||||
| Subcellular location |
Endoplasmic reticulum membrane
|
|||||
| Function | May play a role in anterograde transport of membrane proteins from the endoplasmic reticulum to the Golgi. May be involved in CASP8-mediated apoptosis. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
TH211 Probe Info |
 |
Y90(20.00) | LDD0260 | [2] | |
|
STPyne Probe Info |
 |
K157(1.32); K163(4.63); K96(8.42) | LDD0277 | [3] | |
|
BTD Probe Info |
 |
C171(0.94) | LDD2092 | [4] | |
|
AHL-Pu-1 Probe Info |
 |
C171(2.63) | LDD0168 | [5] | |
|
DBIA Probe Info |
 |
C171(0.97) | LDD1492 | [6] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [7] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [7] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [7] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [8] | |
|
IPM Probe Info |
 |
N.A. | LDD0025 | [9] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [9] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [10] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [9] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [11] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [11] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [11] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [12] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [10] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C082 Probe Info |
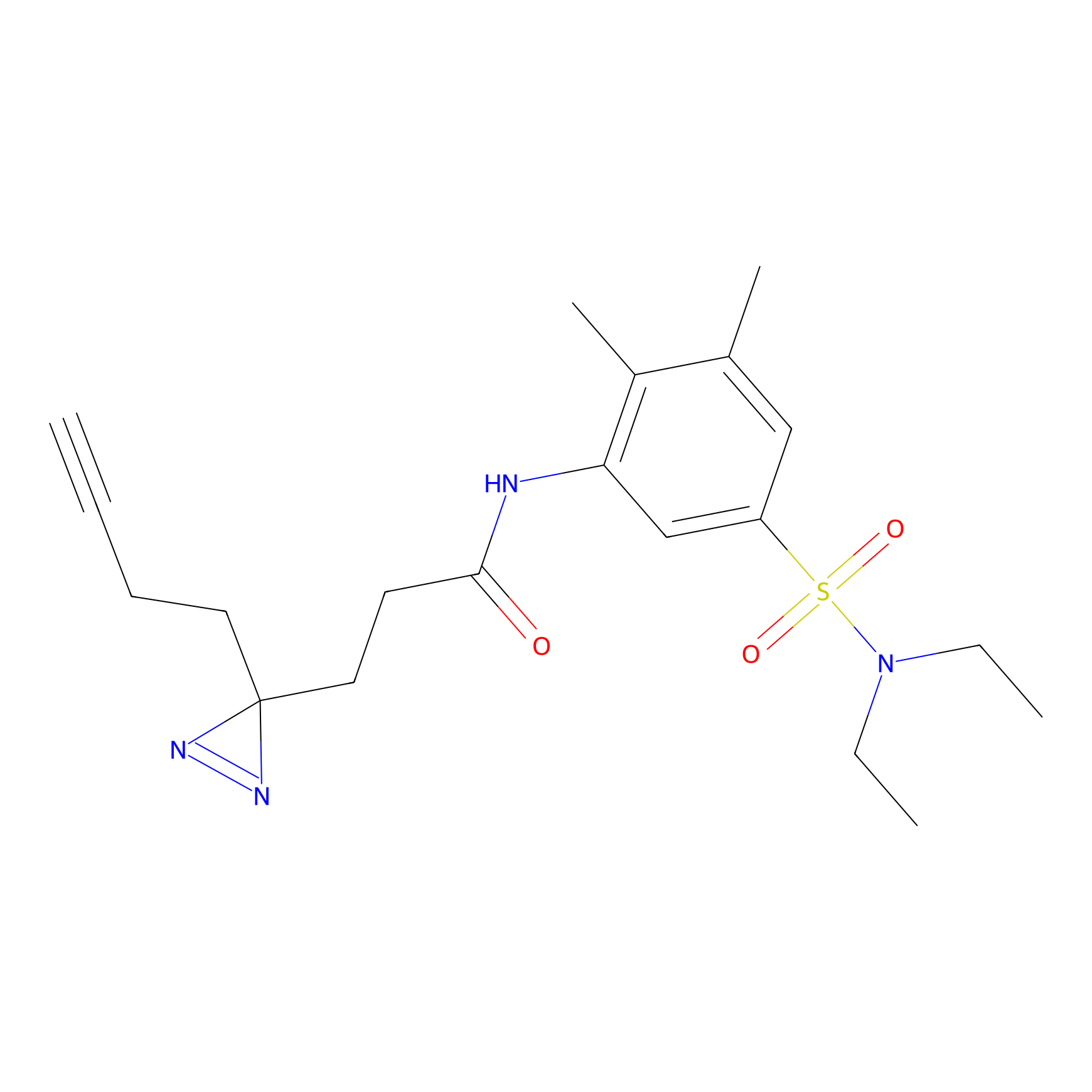 |
6.68 | LDD1774 | [13] | |
|
C083 Probe Info |
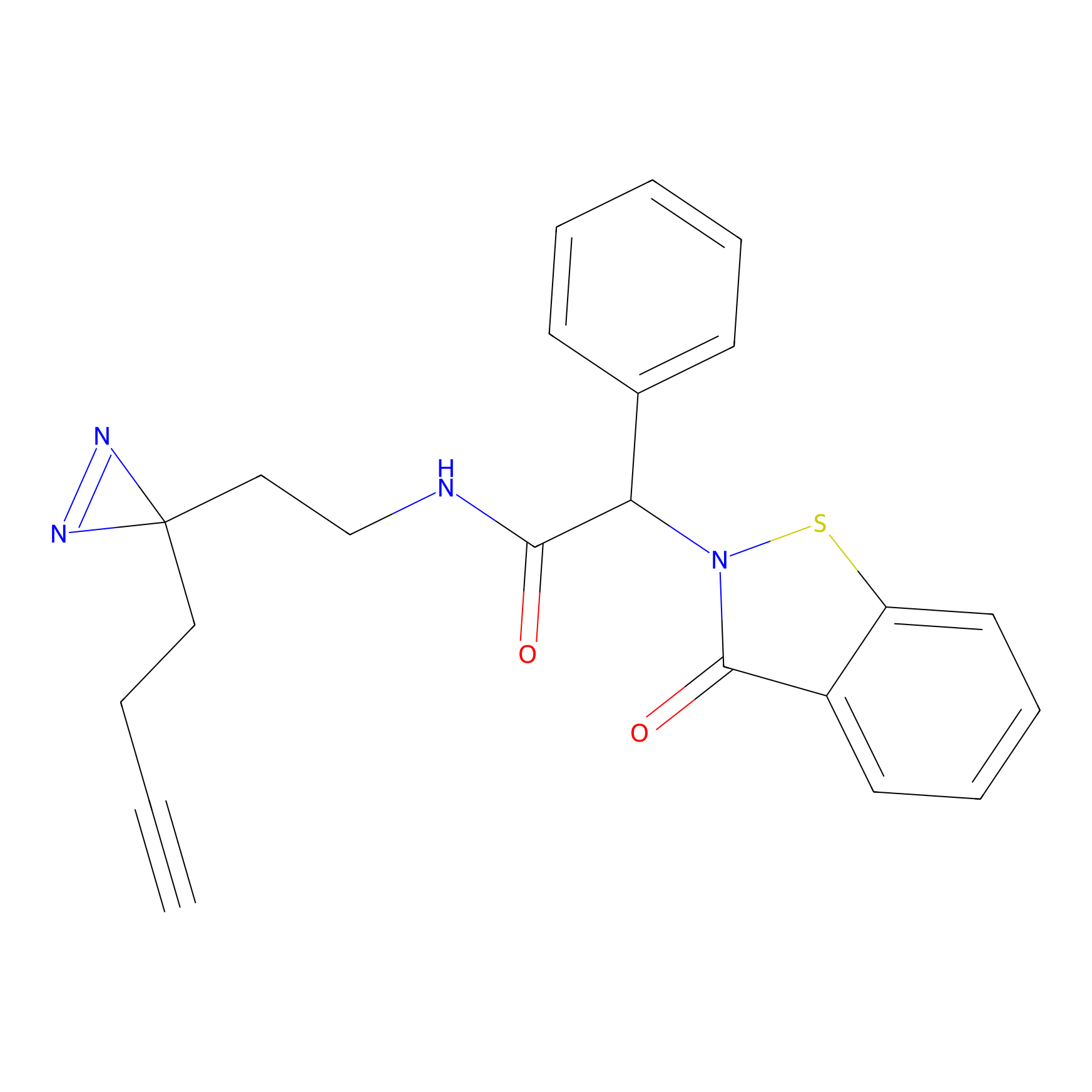 |
7.78 | LDD1775 | [13] | |
|
C087 Probe Info |
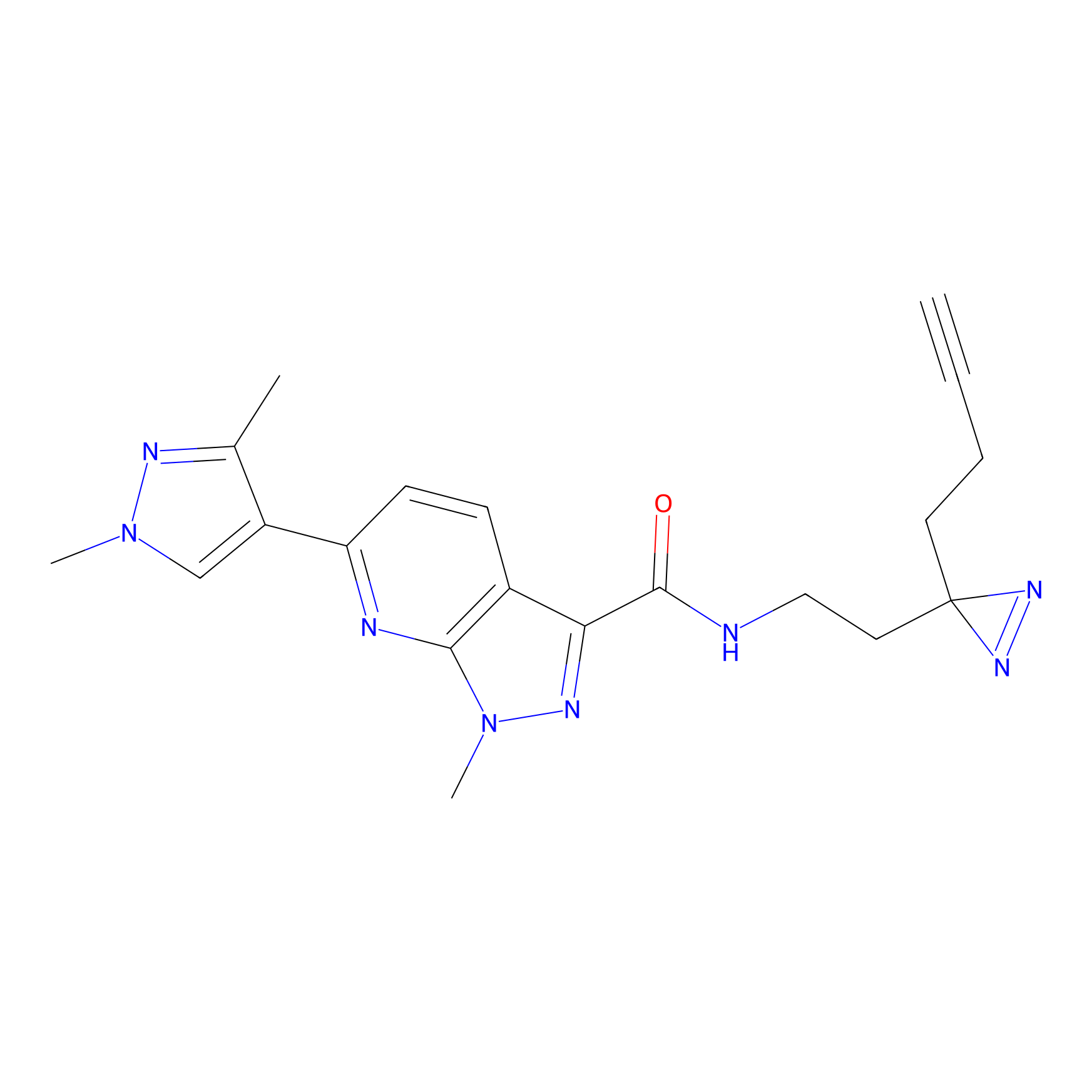 |
47.84 | LDD1779 | [13] | |
|
C111 Probe Info |
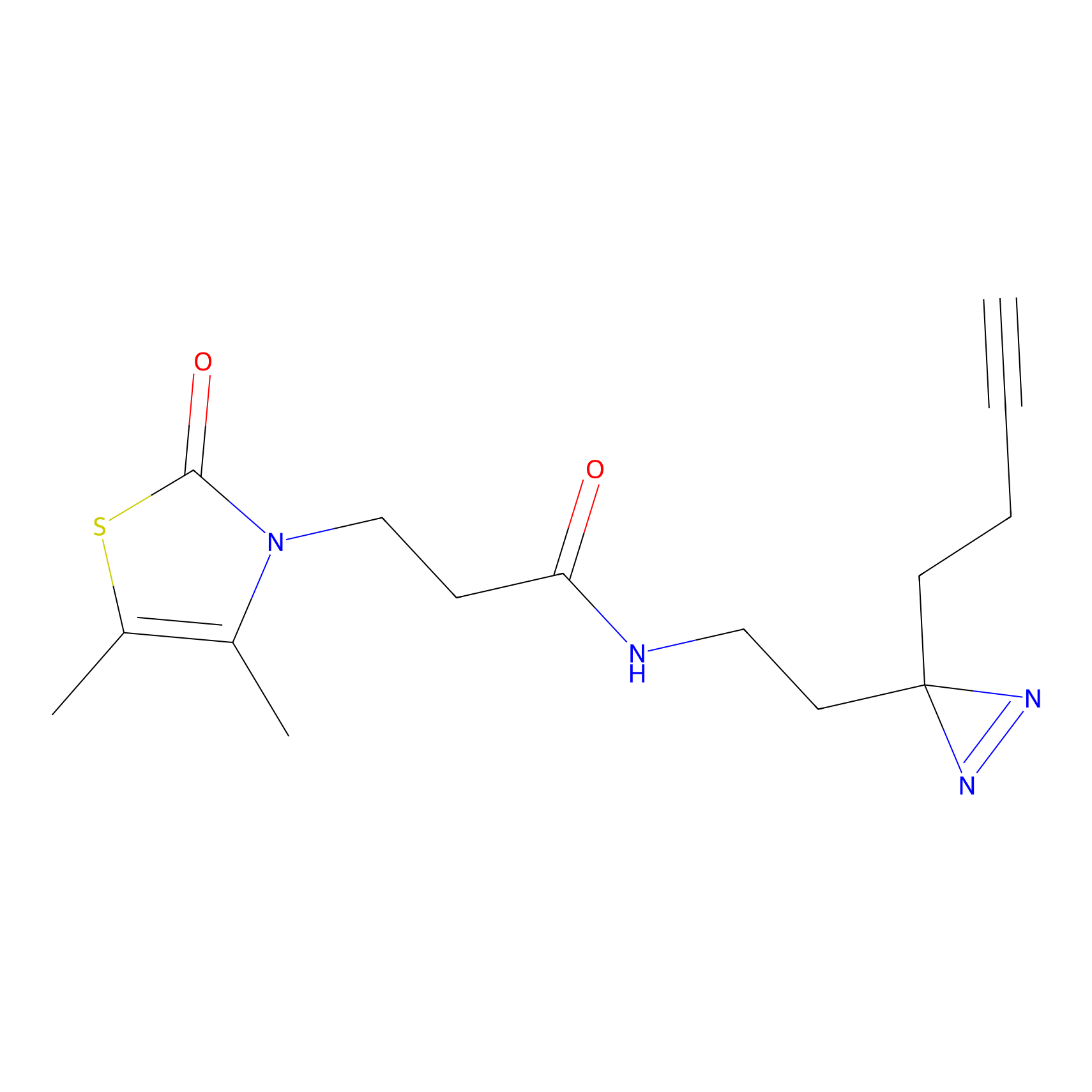 |
6.96 | LDD1798 | [13] | |
|
C112 Probe Info |
 |
20.68 | LDD1799 | [13] | |
|
C158 Probe Info |
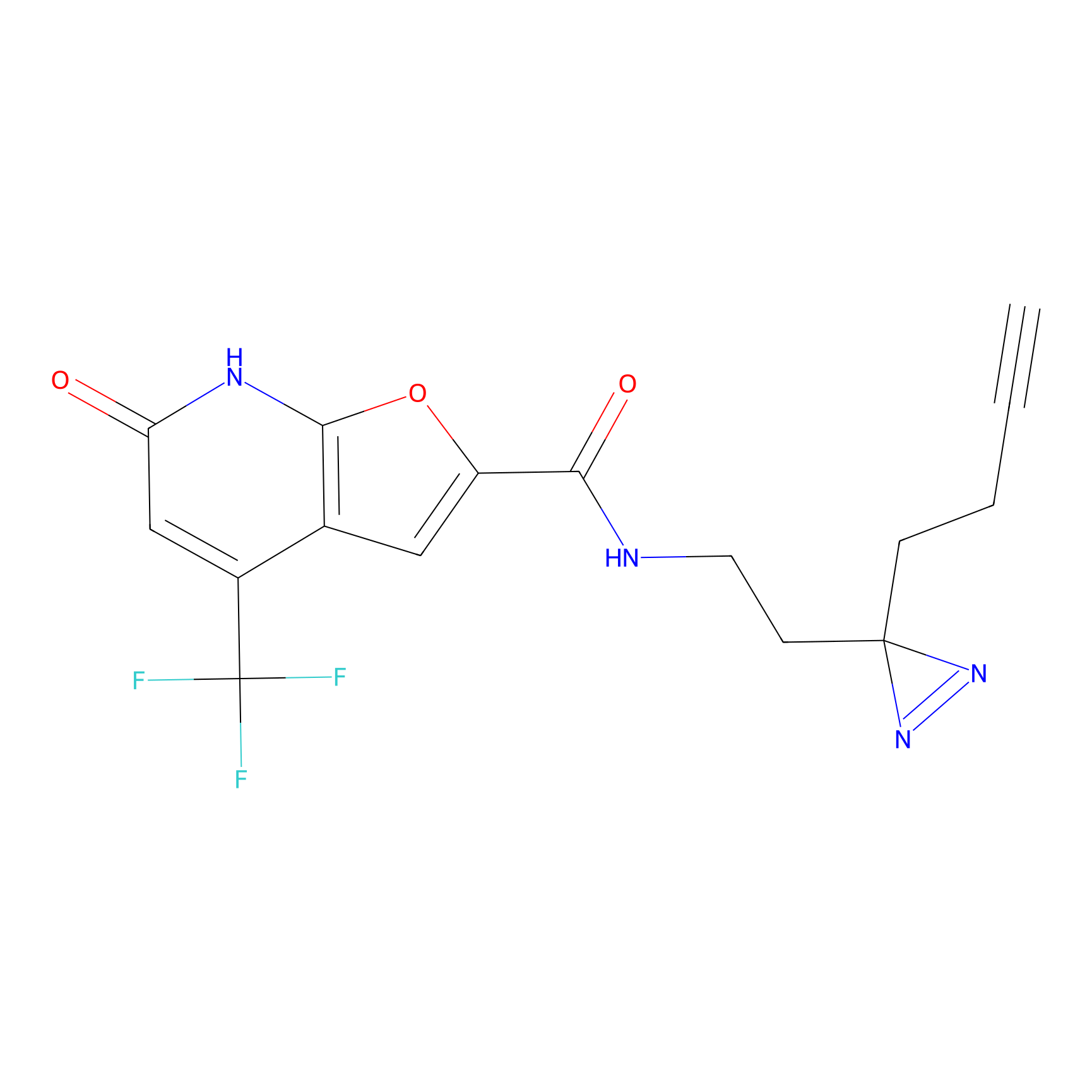 |
11.71 | LDD1838 | [13] | |
|
C169 Probe Info |
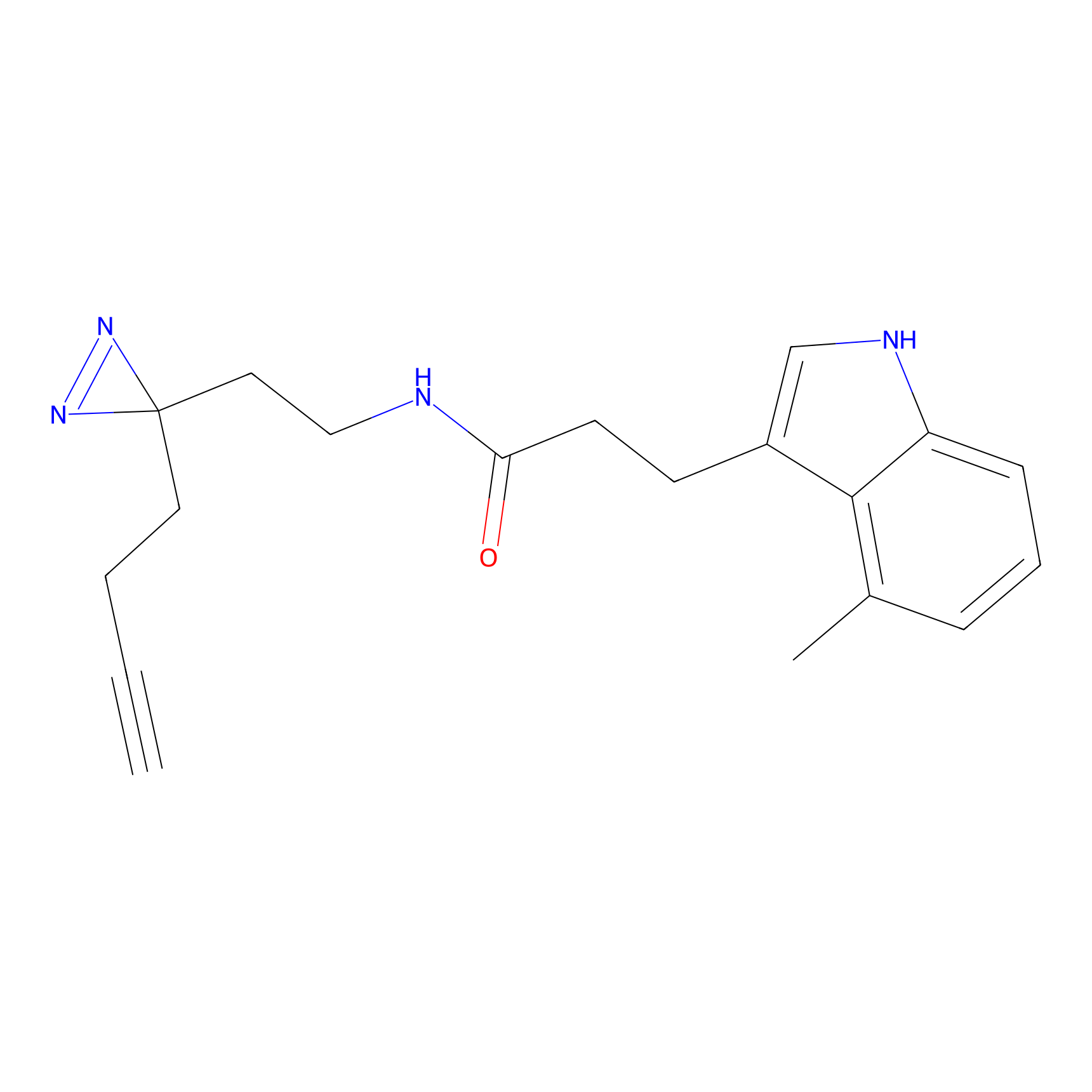 |
30.91 | LDD1849 | [13] | |
|
C362 Probe Info |
 |
34.78 | LDD2023 | [13] | |
|
C373 Probe Info |
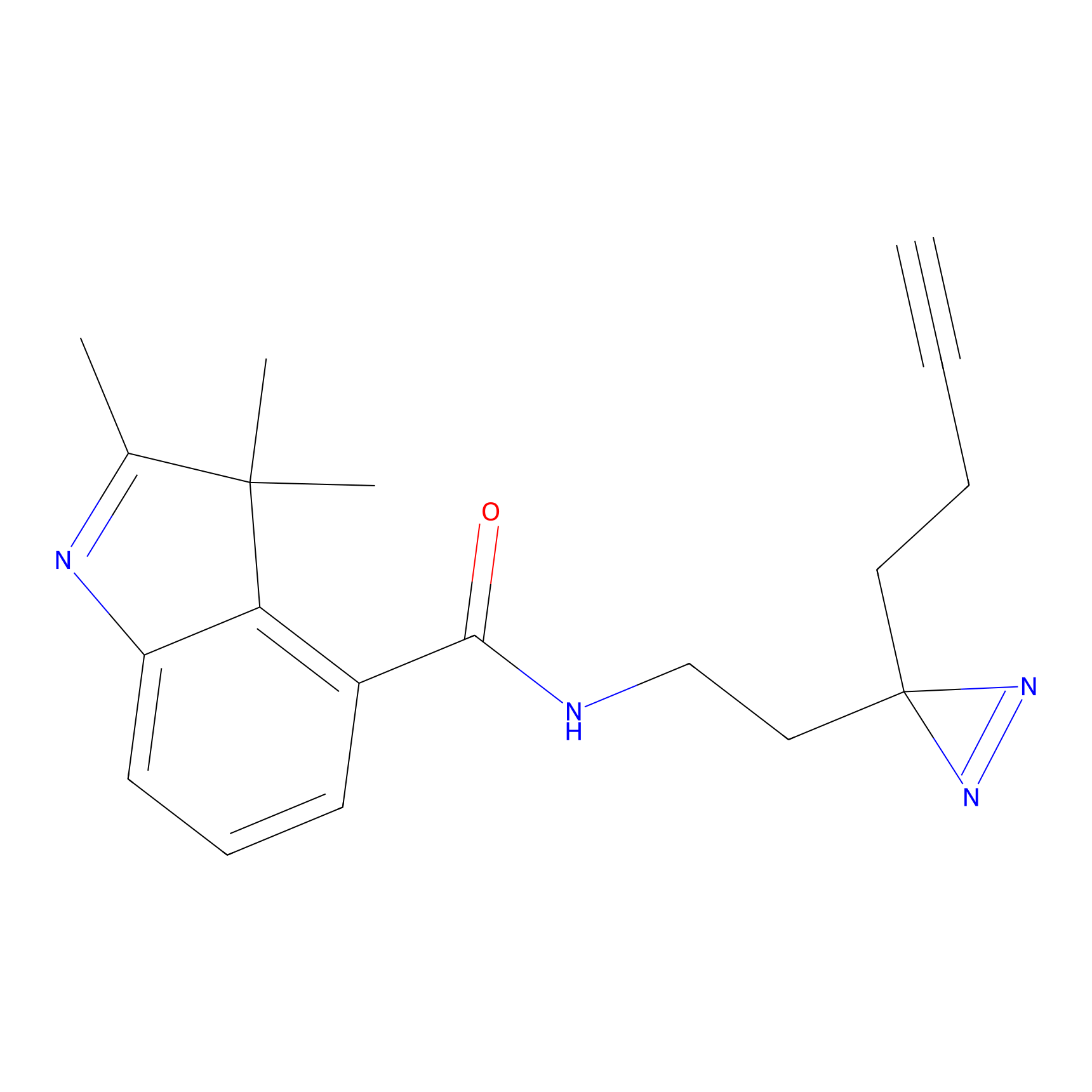 |
6.45 | LDD2033 | [13] | |
|
Dasatinib-CA-8PAP Probe Info |
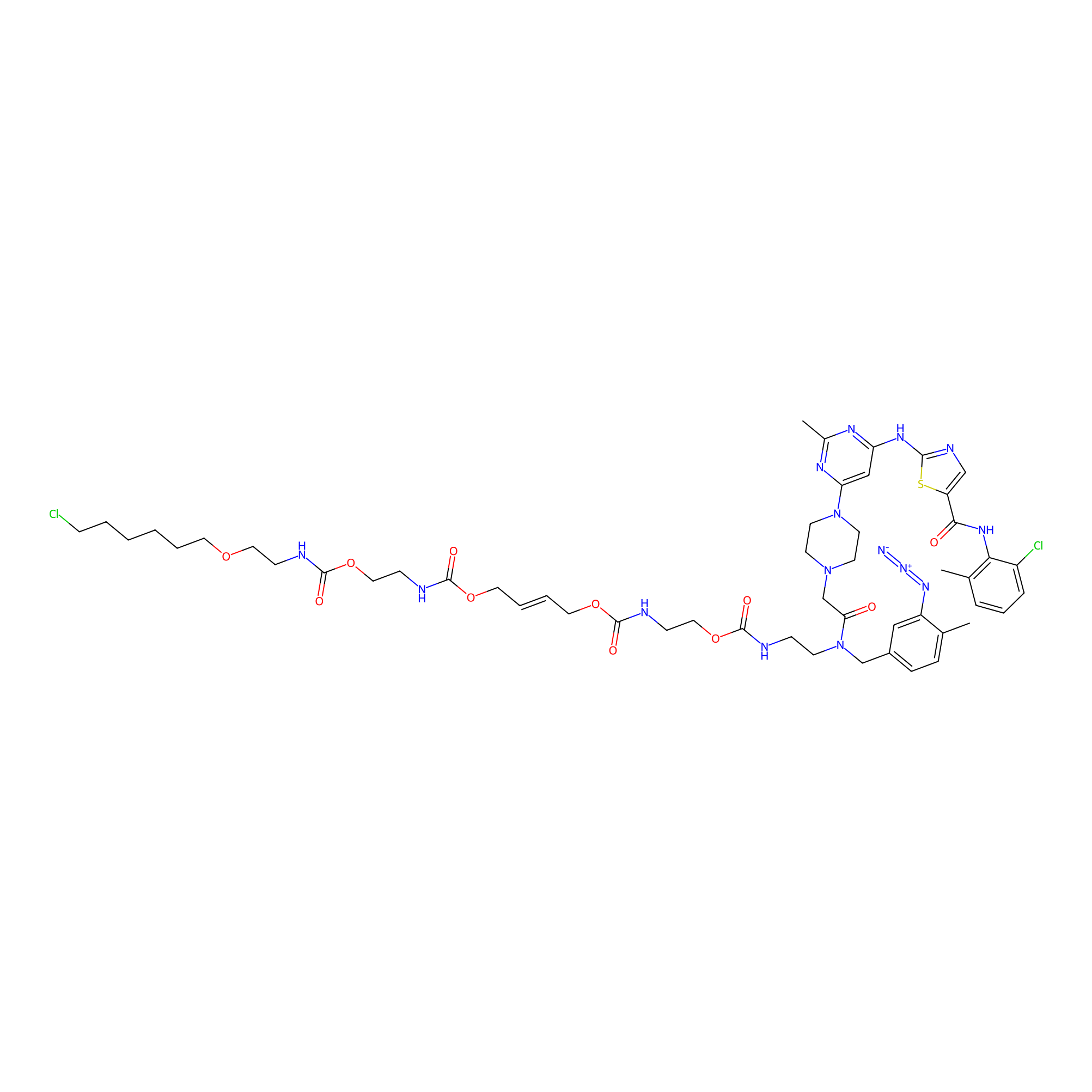 |
8.00 | LDD0364 | [14] | |
|
OEA-DA Probe Info |
 |
20.00 | LDD0046 | [15] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C171(0.81) | LDD2142 | [4] |
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C171(0.95) | LDD2112 | [4] |
| LDCM0502 | 1-(Cyanoacetyl)piperidine | MDA-MB-231 | C171(0.64) | LDD2095 | [4] |
| LDCM0025 | 4SU-RNA | HEK-293T | C171(2.63) | LDD0168 | [5] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C171(2.30) | LDD0169 | [5] |
| LDCM0156 | Aniline | NCI-H1299 | 15.00 | LDD0403 | [1] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [11] |
| LDCM0632 | CL-Sc | Hep-G2 | C171(0.82); C171(0.49) | LDD2227 | [10] |
| LDCM0097 | Dasatinib | K562 | 8.00 | LDD0364 | [14] |
| LDCM0625 | F8 | Ramos | C171(5.99) | LDD2187 | [16] |
| LDCM0573 | Fragment11 | Ramos | C171(12.06) | LDD2190 | [16] |
| LDCM0576 | Fragment14 | Ramos | C171(0.55) | LDD2193 | [16] |
| LDCM0586 | Fragment28 | Ramos | C171(1.09) | LDD2198 | [16] |
| LDCM0566 | Fragment4 | Ramos | C171(0.87) | LDD2184 | [16] |
| LDCM0569 | Fragment7 | Ramos | C171(0.99) | LDD2186 | [16] |
| LDCM0022 | KB02 | HEK-293T | C171(0.97) | LDD1492 | [6] |
| LDCM0023 | KB03 | HEK-293T | C171(1.01) | LDD1497 | [6] |
| LDCM0024 | KB05 | HEK-293T | C171(0.87) | LDD1502 | [6] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [11] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C171(0.94) | LDD2092 | [4] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C171(0.70) | LDD2100 | [4] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C171(0.82) | LDD2104 | [4] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C171(0.55) | LDD2107 | [4] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C171(0.82) | LDD2108 | [4] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C171(0.91) | LDD2114 | [4] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C171(1.08) | LDD2137 | [4] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C171(0.79) | LDD2141 | [4] |
| LDCM0551 | Nucleophilic fragment 5b | MDA-MB-231 | C171(1.62) | LDD2145 | [4] |
References
