Details of the Target
General Information of Target
| Target ID | LDTP13204 | |||||
|---|---|---|---|---|---|---|
| Target Name | Gamma-adducin (ADD3) | |||||
| Gene Name | ADD3 | |||||
| Gene ID | 120 | |||||
| Synonyms |
ADDL; Gamma-adducin; Adducin-like protein 70 |
|||||
| 3D Structure | ||||||
| Sequence |
MAEPSGSPVHVQLPQQAAPVTAAAAAAPAAATAAPAPAAPAAPAPAPAPAAQAVGWPICR
DAYELQEVIGSGATAVVQAALCKPRQERVAIKRINLEKCQTSMDELLKEIQAMSQCSHPN VVTYYTSFVVKDELWLVMKLLSGGSMLDIIKYIVNRGEHKNGVLEEAIIATILKEVLEGL DYLHRNGQIHRDLKAGNILLGEDGSVQIADFGVSAFLATGGDVTRNKVRKTFVGTPCWMA PEVMEQVRGYDFKADMWSFGITAIELATGAAPYHKYPPMKVLMLTLQNDPPTLETGVEDK EMMKKYGKSFRKLLSLCLQKDPSKRPTAAELLKCKFFQKAKNREYLIEKLLTRTPDIAQR AKKVRRVPGSSGHLHKTEDGDWEWSDDEMDEKSEEGKAAFSQEKSRRVKEENPEIAVSAS TIPEQIQSLSVHDSQGPPNANEDYREASSCAVNLVLRLRNSRKELNDIRFEFTPGRDTAD GVSQELFSAGLVDGHDVVIVAANLQKIVDDPKALKTLTFKLASGCDGSEIPDEVKLIGFA QLSVS |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Aldolase class II family, Adducin subfamily
|
|||||
| Subcellular location |
Cytoplasm, cytoskeleton
|
|||||
| Function |
Membrane-cytoskeleton-associated protein that promotes the assembly of the spectrin-actin network. Plays a role in actin filament capping. Binds to calmodulin (Probable). Involved in myogenic reactivity of the renal afferent arteriole (Af-art), renal interlobular arteries and middle cerebral artery (MCA) to increased perfusion pressure. Involved in regulation of potassium channels in the vascular smooth muscle cells (VSMCs) of the Af-art and MCA ex vivo. Involved in regulation of glomerular capillary pressure, glomerular filtration rate (GFR) and glomerular nephrin expression in response to hypertension. Involved in renal blood flow (RBF) autoregulation. Plays a role in podocyte structure and function. Regulates globular monomer actin (G-actin) and filamentous polymer actin (F-actin) ratios in the primary podocytes affecting actin cytoskeleton organization. Regulates expression of synaptopodin, RhoA, Rac1 and CDC42 in the renal cortex and the primary podocytes. Regulates expression of nephrin in the glomeruli and in the primary podocytes, expression of nephrin and podocinin in the renal cortex, and expression of focal adhesion proteins integrin alpha-3 and integrin beta-1 in the glomeruli. Involved in cell migration and cell adhesion of podocytes, and in podocyte foot process effacement. Regulates expression of profibrotics markers MMP2, MMP9, TGF beta-1, tubular tight junction protein E-cadherin, and mesenchymal markers vimentin and alpha-SMA. Promotes the growth of neurites.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| CAL62 | SNV: p.P85L | DBIA Probe Info | |||
| DU145 | SNV: p.S474L; p.T653I | DBIA Probe Info | |||
| MCC13 | SNV: p.K397Q | DBIA Probe Info | |||
| NALM6 | SNV: p.R138H | DBIA Probe Info | |||
| OPM2 | SNV: p.E623V | DBIA Probe Info | |||
| RKO | SNV: p.L111R | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K465(3.22); K470(1.10) | LDD0277 | [1] | |
|
Probe 1 Probe Info |
 |
Y35(13.43) | LDD3495 | [2] | |
|
BTD Probe Info |
 |
C286(0.84) | LDD2093 | [3] | |
|
AHL-Pu-1 Probe Info |
 |
C73(2.31) | LDD0168 | [4] | |
|
DBIA Probe Info |
 |
C73(1.07) | LDD0080 | [5] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0358 | [6] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C73(0.00); C286(0.00); C245(0.00) | LDD0038 | [7] | |
|
IA-alkyne Probe Info |
 |
C73(0.00); C286(0.00) | LDD0036 | [7] | |
|
Lodoacetamide azide Probe Info |
 |
C245(0.00); C286(0.00); C73(0.00) | LDD0037 | [7] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2225 | [8] | |
|
IPM Probe Info |
 |
N.A. | LDD2156 | [9] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [10] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [11] | |
|
Phosphinate-6 Probe Info |
 |
C139(0.00); C286(0.00) | LDD0018 | [12] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [6] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [13] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [13] | |
|
AOyne Probe Info |
 |
13.70 | LDD0443 | [14] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C040 Probe Info |
 |
10.41 | LDD1740 | [15] | |
|
C145 Probe Info |
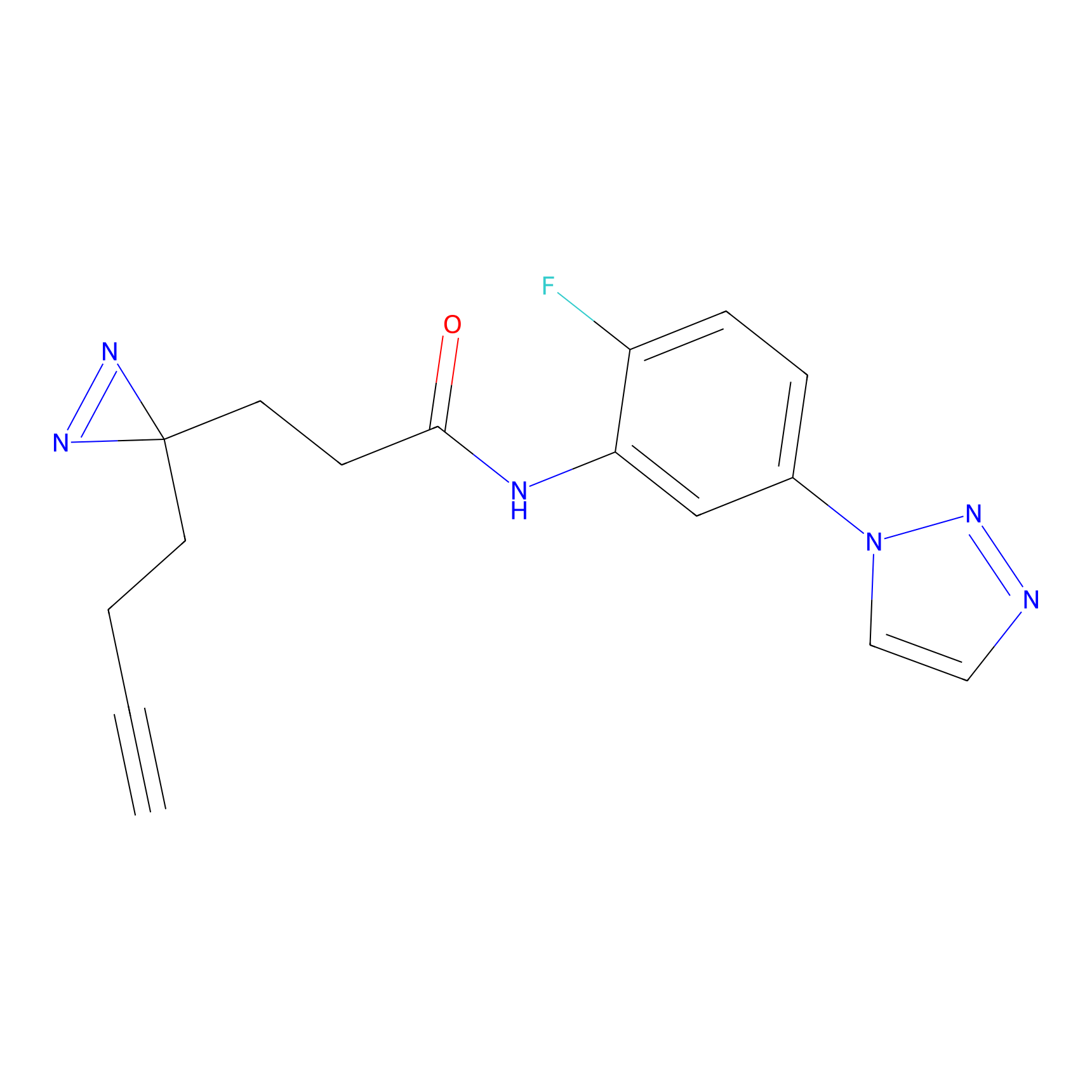 |
9.71 | LDD1827 | [15] | |
|
C382 Probe Info |
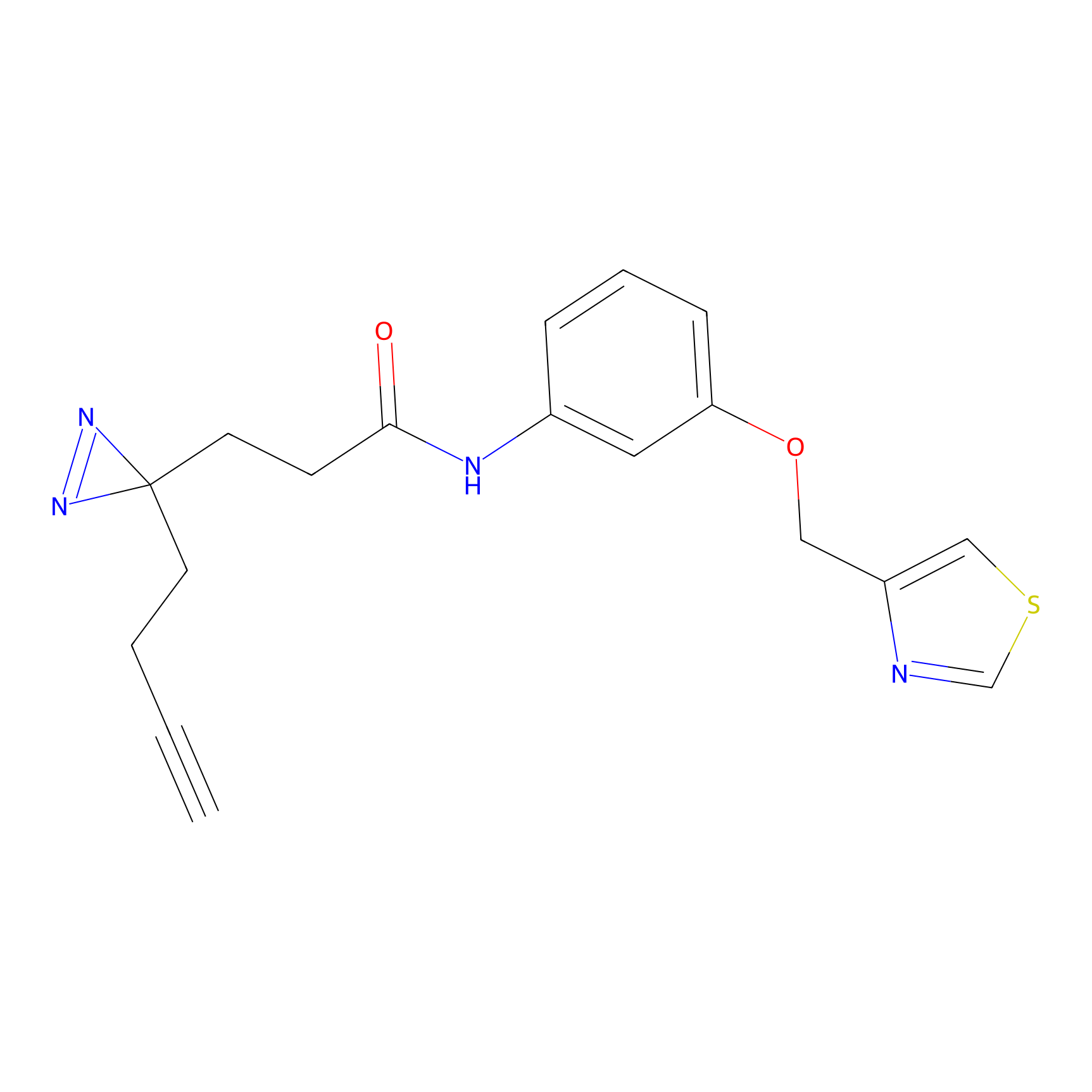 |
9.78 | LDD2041 | [15] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C286(0.89) | LDD2152 | [3] |
| LDCM0025 | 4SU-RNA | HEK-293T | C73(2.31) | LDD0168 | [4] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C73(2.90) | LDD0169 | [4] |
| LDCM0214 | AC1 | HEK-293T | C286(0.91); C73(0.86); C245(0.99) | LDD1507 | [16] |
| LDCM0215 | AC10 | HCT 116 | C286(0.98) | LDD0532 | [5] |
| LDCM0226 | AC11 | HCT 116 | C286(0.96) | LDD0543 | [5] |
| LDCM0230 | AC113 | PaTu 8988t | C286(1.11) | LDD1109 | [5] |
| LDCM0231 | AC114 | PaTu 8988t | C286(1.08) | LDD1110 | [5] |
| LDCM0232 | AC115 | PaTu 8988t | C286(0.97) | LDD1111 | [5] |
| LDCM0233 | AC116 | PaTu 8988t | C286(1.18) | LDD1112 | [5] |
| LDCM0234 | AC117 | PaTu 8988t | C286(1.08) | LDD1113 | [5] |
| LDCM0235 | AC118 | PaTu 8988t | C286(1.00) | LDD1114 | [5] |
| LDCM0236 | AC119 | PaTu 8988t | C286(1.04) | LDD1115 | [5] |
| LDCM0237 | AC12 | HCT 116 | C286(0.98) | LDD0554 | [5] |
| LDCM0238 | AC120 | PaTu 8988t | C286(1.04) | LDD1117 | [5] |
| LDCM0239 | AC121 | PaTu 8988t | C286(1.11) | LDD1118 | [5] |
| LDCM0240 | AC122 | PaTu 8988t | C286(1.09) | LDD1119 | [5] |
| LDCM0241 | AC123 | PaTu 8988t | C286(0.94) | LDD1120 | [5] |
| LDCM0242 | AC124 | PaTu 8988t | C286(1.08) | LDD1121 | [5] |
| LDCM0243 | AC125 | PaTu 8988t | C286(1.02) | LDD1122 | [5] |
| LDCM0244 | AC126 | PaTu 8988t | C286(1.13) | LDD1123 | [5] |
| LDCM0245 | AC127 | PaTu 8988t | C286(1.00) | LDD1124 | [5] |
| LDCM0246 | AC128 | PaTu 8988t | C286(1.03) | LDD1125 | [5] |
| LDCM0247 | AC129 | PaTu 8988t | C286(1.01) | LDD1126 | [5] |
| LDCM0249 | AC130 | PaTu 8988t | C286(0.99) | LDD1128 | [5] |
| LDCM0250 | AC131 | PaTu 8988t | C286(0.92) | LDD1129 | [5] |
| LDCM0251 | AC132 | PaTu 8988t | C286(1.04) | LDD1130 | [5] |
| LDCM0252 | AC133 | PaTu 8988t | C286(1.23) | LDD1131 | [5] |
| LDCM0253 | AC134 | PaTu 8988t | C286(1.11) | LDD1132 | [5] |
| LDCM0254 | AC135 | PaTu 8988t | C286(1.12) | LDD1133 | [5] |
| LDCM0255 | AC136 | PaTu 8988t | C286(1.11) | LDD1134 | [5] |
| LDCM0256 | AC137 | PaTu 8988t | C286(1.20) | LDD1135 | [5] |
| LDCM0257 | AC138 | PaTu 8988t | C286(0.97) | LDD1136 | [5] |
| LDCM0258 | AC139 | PaTu 8988t | C286(0.98) | LDD1137 | [5] |
| LDCM0259 | AC14 | HCT 116 | C286(1.03) | LDD0576 | [5] |
| LDCM0260 | AC140 | PaTu 8988t | C286(0.95) | LDD1139 | [5] |
| LDCM0261 | AC141 | PaTu 8988t | C286(0.91) | LDD1140 | [5] |
| LDCM0262 | AC142 | PaTu 8988t | C286(1.07) | LDD1141 | [5] |
| LDCM0263 | AC143 | HCT 116 | C286(0.94) | LDD0580 | [5] |
| LDCM0264 | AC144 | HCT 116 | C286(0.85) | LDD0581 | [5] |
| LDCM0265 | AC145 | HCT 116 | C286(0.87) | LDD0582 | [5] |
| LDCM0266 | AC146 | HCT 116 | C286(0.89) | LDD0583 | [5] |
| LDCM0267 | AC147 | HCT 116 | C286(0.93) | LDD0584 | [5] |
| LDCM0268 | AC148 | HCT 116 | C286(1.12) | LDD0585 | [5] |
| LDCM0269 | AC149 | HCT 116 | C286(1.04) | LDD0586 | [5] |
| LDCM0270 | AC15 | HCT 116 | C286(1.02) | LDD0587 | [5] |
| LDCM0271 | AC150 | HCT 116 | C286(0.90) | LDD0588 | [5] |
| LDCM0272 | AC151 | HCT 116 | C286(0.80) | LDD0589 | [5] |
| LDCM0273 | AC152 | HCT 116 | C286(0.93) | LDD0590 | [5] |
| LDCM0274 | AC153 | HCT 116 | C286(1.14) | LDD0591 | [5] |
| LDCM0621 | AC154 | HCT 116 | C286(0.92) | LDD2158 | [5] |
| LDCM0622 | AC155 | HCT 116 | C286(0.85) | LDD2159 | [5] |
| LDCM0623 | AC156 | HCT 116 | C286(0.90) | LDD2160 | [5] |
| LDCM0624 | AC157 | HCT 116 | C286(0.86) | LDD2161 | [5] |
| LDCM0276 | AC17 | HEK-293T | C286(0.94); C73(0.98); C245(1.16) | LDD1515 | [16] |
| LDCM0277 | AC18 | HEK-293T | C286(0.99); C73(1.00); C245(0.89) | LDD1516 | [16] |
| LDCM0278 | AC19 | HEK-293T | C286(0.96); C73(0.94); C245(0.86) | LDD1517 | [16] |
| LDCM0279 | AC2 | HEK-293T | C286(0.98); C73(1.03); C245(0.90) | LDD1518 | [16] |
| LDCM0280 | AC20 | HEK-293T | C286(1.00); C73(1.06); C245(1.11) | LDD1519 | [16] |
| LDCM0281 | AC21 | HEK-293T | C286(1.00); C73(1.03); C245(0.98) | LDD1520 | [16] |
| LDCM0282 | AC22 | HEK-293T | C286(0.94); C73(1.01) | LDD1521 | [16] |
| LDCM0283 | AC23 | HEK-293T | C286(0.98); C73(1.03); C245(1.08) | LDD1522 | [16] |
| LDCM0284 | AC24 | HEK-293T | C286(1.03); C73(0.98); C245(1.07) | LDD1523 | [16] |
| LDCM0285 | AC25 | HCT 116 | C286(0.98) | LDD0602 | [5] |
| LDCM0286 | AC26 | HCT 116 | C286(0.89) | LDD0603 | [5] |
| LDCM0287 | AC27 | HCT 116 | C286(0.92) | LDD0604 | [5] |
| LDCM0288 | AC28 | HCT 116 | C286(0.93) | LDD0605 | [5] |
| LDCM0289 | AC29 | HCT 116 | C286(0.91) | LDD0606 | [5] |
| LDCM0290 | AC3 | HEK-293T | C286(0.88); C73(1.02); C245(1.21) | LDD1529 | [16] |
| LDCM0291 | AC30 | HCT 116 | C286(0.91) | LDD0608 | [5] |
| LDCM0292 | AC31 | HCT 116 | C286(0.93) | LDD0609 | [5] |
| LDCM0293 | AC32 | HCT 116 | C286(0.95) | LDD0610 | [5] |
| LDCM0294 | AC33 | HCT 116 | C286(0.89) | LDD0611 | [5] |
| LDCM0295 | AC34 | HCT 116 | C286(0.92) | LDD0612 | [5] |
| LDCM0296 | AC35 | HCT 116 | C286(1.01) | LDD0613 | [5] |
| LDCM0297 | AC36 | HCT 116 | C286(1.00) | LDD0614 | [5] |
| LDCM0298 | AC37 | HCT 116 | C286(0.96) | LDD0615 | [5] |
| LDCM0299 | AC38 | HCT 116 | C286(0.89) | LDD0616 | [5] |
| LDCM0300 | AC39 | HCT 116 | C286(1.08) | LDD0617 | [5] |
| LDCM0301 | AC4 | HEK-293T | C286(1.01); C73(1.00); C245(1.01) | LDD1540 | [16] |
| LDCM0302 | AC40 | HCT 116 | C286(1.05) | LDD0619 | [5] |
| LDCM0303 | AC41 | HCT 116 | C286(1.01) | LDD0620 | [5] |
| LDCM0304 | AC42 | HCT 116 | C286(0.98) | LDD0621 | [5] |
| LDCM0305 | AC43 | HCT 116 | C286(0.95) | LDD0622 | [5] |
| LDCM0306 | AC44 | HCT 116 | C286(1.03) | LDD0623 | [5] |
| LDCM0307 | AC45 | HCT 116 | C286(1.08) | LDD0624 | [5] |
| LDCM0308 | AC46 | HCT 116 | C286(0.97) | LDD0625 | [5] |
| LDCM0309 | AC47 | HCT 116 | C286(1.14) | LDD0626 | [5] |
| LDCM0310 | AC48 | HCT 116 | C286(1.14) | LDD0627 | [5] |
| LDCM0311 | AC49 | HCT 116 | C286(1.24) | LDD0628 | [5] |
| LDCM0312 | AC5 | HEK-293T | C286(1.06); C73(1.00); C245(1.06) | LDD1551 | [16] |
| LDCM0313 | AC50 | HCT 116 | C286(1.09) | LDD0630 | [5] |
| LDCM0314 | AC51 | HCT 116 | C286(1.19) | LDD0631 | [5] |
| LDCM0315 | AC52 | HCT 116 | C286(1.09) | LDD0632 | [5] |
| LDCM0316 | AC53 | HCT 116 | C286(1.01) | LDD0633 | [5] |
| LDCM0317 | AC54 | HCT 116 | C286(1.07) | LDD0634 | [5] |
| LDCM0318 | AC55 | HCT 116 | C286(1.11) | LDD0635 | [5] |
| LDCM0319 | AC56 | HCT 116 | C286(1.34) | LDD0636 | [5] |
| LDCM0320 | AC57 | HCT 116 | C286(1.07) | LDD0637 | [5] |
| LDCM0321 | AC58 | HCT 116 | C286(1.03) | LDD0638 | [5] |
| LDCM0322 | AC59 | HCT 116 | C286(1.14) | LDD0639 | [5] |
| LDCM0323 | AC6 | HCT 116 | C286(0.97) | LDD0640 | [5] |
| LDCM0324 | AC60 | HCT 116 | C286(1.02) | LDD0641 | [5] |
| LDCM0325 | AC61 | HCT 116 | C286(0.96) | LDD0642 | [5] |
| LDCM0326 | AC62 | HCT 116 | C286(0.97) | LDD0643 | [5] |
| LDCM0327 | AC63 | HCT 116 | C286(1.00) | LDD0644 | [5] |
| LDCM0328 | AC64 | HCT 116 | C286(1.32) | LDD0645 | [5] |
| LDCM0329 | AC65 | HCT 116 | C286(1.35) | LDD0646 | [5] |
| LDCM0330 | AC66 | HCT 116 | C286(1.38) | LDD0647 | [5] |
| LDCM0331 | AC67 | HCT 116 | C286(1.02) | LDD0648 | [5] |
| LDCM0332 | AC68 | PaTu 8988t | C73(0.98) | LDD1211 | [5] |
| LDCM0333 | AC69 | PaTu 8988t | C73(1.12) | LDD1212 | [5] |
| LDCM0334 | AC7 | HCT 116 | C286(0.98) | LDD0651 | [5] |
| LDCM0335 | AC70 | PaTu 8988t | C73(1.12) | LDD1214 | [5] |
| LDCM0336 | AC71 | PaTu 8988t | C73(1.09) | LDD1215 | [5] |
| LDCM0337 | AC72 | PaTu 8988t | C73(1.05) | LDD1216 | [5] |
| LDCM0338 | AC73 | PaTu 8988t | C73(0.95) | LDD1217 | [5] |
| LDCM0339 | AC74 | PaTu 8988t | C73(1.15) | LDD1218 | [5] |
| LDCM0340 | AC75 | PaTu 8988t | C73(1.19) | LDD1219 | [5] |
| LDCM0341 | AC76 | PaTu 8988t | C73(1.04) | LDD1220 | [5] |
| LDCM0342 | AC77 | PaTu 8988t | C73(0.95) | LDD1221 | [5] |
| LDCM0343 | AC78 | PaTu 8988t | C73(1.17) | LDD1222 | [5] |
| LDCM0344 | AC79 | PaTu 8988t | C73(1.21) | LDD1223 | [5] |
| LDCM0345 | AC8 | HCT 116 | C286(0.96) | LDD0662 | [5] |
| LDCM0346 | AC80 | PaTu 8988t | C73(1.14) | LDD1225 | [5] |
| LDCM0347 | AC81 | PaTu 8988t | C73(1.22) | LDD1226 | [5] |
| LDCM0348 | AC82 | PaTu 8988t | C73(1.35) | LDD1227 | [5] |
| LDCM0349 | AC83 | HCT 116 | C286(1.07) | LDD0666 | [5] |
| LDCM0350 | AC84 | HCT 116 | C286(1.07) | LDD0667 | [5] |
| LDCM0351 | AC85 | HCT 116 | C286(1.06) | LDD0668 | [5] |
| LDCM0352 | AC86 | HCT 116 | C286(1.00) | LDD0669 | [5] |
| LDCM0353 | AC87 | HCT 116 | C286(1.00) | LDD0670 | [5] |
| LDCM0354 | AC88 | HCT 116 | C286(0.92) | LDD0671 | [5] |
| LDCM0355 | AC89 | HCT 116 | C286(1.05) | LDD0672 | [5] |
| LDCM0357 | AC90 | HCT 116 | C286(0.90) | LDD0674 | [5] |
| LDCM0358 | AC91 | HCT 116 | C286(1.17) | LDD0675 | [5] |
| LDCM0359 | AC92 | HCT 116 | C286(1.09) | LDD0676 | [5] |
| LDCM0360 | AC93 | HCT 116 | C286(0.90) | LDD0677 | [5] |
| LDCM0361 | AC94 | HCT 116 | C286(1.20) | LDD0678 | [5] |
| LDCM0362 | AC95 | HCT 116 | C286(1.02) | LDD0679 | [5] |
| LDCM0363 | AC96 | HCT 116 | C286(1.05) | LDD0680 | [5] |
| LDCM0364 | AC97 | HCT 116 | C286(0.96) | LDD0681 | [5] |
| LDCM0248 | AKOS034007472 | HCT 116 | C286(1.02) | LDD0565 | [5] |
| LDCM0356 | AKOS034007680 | HCT 116 | C286(0.96) | LDD0673 | [5] |
| LDCM0275 | AKOS034007705 | HCT 116 | C286(1.17) | LDD0592 | [5] |
| LDCM0108 | Chloroacetamide | HeLa | C286(0.00); C73(0.00) | LDD0222 | [13] |
| LDCM0367 | CL1 | PaTu 8988t | C73(0.94); C286(0.85) | LDD1246 | [5] |
| LDCM0368 | CL10 | PaTu 8988t | C73(1.35); C286(0.81) | LDD1247 | [5] |
| LDCM0369 | CL100 | HEK-293T | C286(0.97); C73(1.00); C245(1.00) | LDD1573 | [16] |
| LDCM0370 | CL101 | HCT 116 | C286(1.01) | LDD0687 | [5] |
| LDCM0371 | CL102 | HCT 116 | C286(0.98) | LDD0688 | [5] |
| LDCM0372 | CL103 | HCT 116 | C286(0.92) | LDD0689 | [5] |
| LDCM0373 | CL104 | HCT 116 | C286(0.94) | LDD0690 | [5] |
| LDCM0374 | CL105 | HEK-293T | C286(0.96); C73(0.92); C245(1.09) | LDD1578 | [16] |
| LDCM0375 | CL106 | HEK-293T | C286(0.99); C73(1.00); C245(1.07) | LDD1579 | [16] |
| LDCM0376 | CL107 | HEK-293T | C286(1.01); C73(1.01); C245(1.09) | LDD1580 | [16] |
| LDCM0377 | CL108 | HEK-293T | C286(0.98); C73(0.99); C245(0.97) | LDD1581 | [16] |
| LDCM0378 | CL109 | HEK-293T | C286(1.03); C73(0.88); C245(1.07) | LDD1582 | [16] |
| LDCM0379 | CL11 | PaTu 8988t | C73(0.91); C286(0.94) | LDD1258 | [5] |
| LDCM0380 | CL110 | HEK-293T | C286(1.05); C73(0.98); C245(0.99) | LDD1584 | [16] |
| LDCM0381 | CL111 | HEK-293T | C286(1.01); C73(0.99); C245(1.10) | LDD1585 | [16] |
| LDCM0382 | CL112 | HCT 116 | C286(0.92) | LDD0699 | [5] |
| LDCM0383 | CL113 | HCT 116 | C286(0.92) | LDD0700 | [5] |
| LDCM0384 | CL114 | HCT 116 | C286(0.89) | LDD0701 | [5] |
| LDCM0385 | CL115 | HCT 116 | C286(0.93) | LDD0702 | [5] |
| LDCM0386 | CL116 | HCT 116 | C286(0.91) | LDD0703 | [5] |
| LDCM0387 | CL117 | HCT 116 | C286(1.19) | LDD0704 | [5] |
| LDCM0388 | CL118 | HCT 116 | C286(1.03) | LDD0705 | [5] |
| LDCM0389 | CL119 | HCT 116 | C286(1.07) | LDD0706 | [5] |
| LDCM0390 | CL12 | PaTu 8988t | C73(1.01); C286(0.88) | LDD1269 | [5] |
| LDCM0391 | CL120 | HCT 116 | C286(1.05) | LDD0708 | [5] |
| LDCM0392 | CL121 | HCT 116 | C286(1.14) | LDD0709 | [5] |
| LDCM0393 | CL122 | HCT 116 | C286(1.02) | LDD0710 | [5] |
| LDCM0394 | CL123 | HCT 116 | C286(1.00) | LDD0711 | [5] |
| LDCM0395 | CL124 | HCT 116 | C286(1.02) | LDD0712 | [5] |
| LDCM0396 | CL125 | HCT 116 | C286(1.05) | LDD0713 | [5] |
| LDCM0397 | CL126 | HCT 116 | C286(0.94) | LDD0714 | [5] |
| LDCM0398 | CL127 | HCT 116 | C286(0.85) | LDD0715 | [5] |
| LDCM0399 | CL128 | HCT 116 | C286(0.88) | LDD0716 | [5] |
| LDCM0400 | CL13 | PaTu 8988t | C73(1.63); C286(0.98) | LDD1279 | [5] |
| LDCM0401 | CL14 | PaTu 8988t | C73(1.66); C286(0.95) | LDD1280 | [5] |
| LDCM0402 | CL15 | PaTu 8988t | C73(1.15); C286(0.92) | LDD1281 | [5] |
| LDCM0403 | CL16 | HCT 116 | C286(1.01) | LDD0720 | [5] |
| LDCM0404 | CL17 | HCT 116 | C286(0.64) | LDD0721 | [5] |
| LDCM0405 | CL18 | HCT 116 | C286(0.94) | LDD0722 | [5] |
| LDCM0406 | CL19 | HCT 116 | C286(0.86) | LDD0723 | [5] |
| LDCM0407 | CL2 | PaTu 8988t | C73(0.98); C286(0.87) | LDD1286 | [5] |
| LDCM0408 | CL20 | HCT 116 | C286(0.82) | LDD0725 | [5] |
| LDCM0409 | CL21 | HCT 116 | C286(0.76) | LDD0726 | [5] |
| LDCM0410 | CL22 | HCT 116 | C286(1.08) | LDD0727 | [5] |
| LDCM0411 | CL23 | HCT 116 | C286(0.91) | LDD0728 | [5] |
| LDCM0412 | CL24 | HCT 116 | C286(0.86) | LDD0729 | [5] |
| LDCM0413 | CL25 | HCT 116 | C286(1.01) | LDD0730 | [5] |
| LDCM0414 | CL26 | HCT 116 | C286(0.85) | LDD0731 | [5] |
| LDCM0415 | CL27 | HCT 116 | C286(0.95) | LDD0732 | [5] |
| LDCM0416 | CL28 | HCT 116 | C286(0.86) | LDD0733 | [5] |
| LDCM0417 | CL29 | HCT 116 | C286(0.91) | LDD0734 | [5] |
| LDCM0418 | CL3 | PaTu 8988t | C73(1.29); C286(0.91) | LDD1297 | [5] |
| LDCM0419 | CL30 | HCT 116 | C286(0.96) | LDD0736 | [5] |
| LDCM0420 | CL31 | PaTu 8988t | C73(1.11); C286(1.00) | LDD1299 | [5] |
| LDCM0421 | CL32 | PaTu 8988t | C73(1.90); C286(1.18) | LDD1300 | [5] |
| LDCM0422 | CL33 | PaTu 8988t | C73(1.59); C286(1.02) | LDD1301 | [5] |
| LDCM0423 | CL34 | PaTu 8988t | C73(1.98); C286(1.04) | LDD1302 | [5] |
| LDCM0424 | CL35 | PaTu 8988t | C286(1.05); C73(1.66) | LDD1303 | [5] |
| LDCM0425 | CL36 | PaTu 8988t | C73(1.27); C286(0.99) | LDD1304 | [5] |
| LDCM0426 | CL37 | PaTu 8988t | C73(2.04); C286(1.27) | LDD1305 | [5] |
| LDCM0428 | CL39 | PaTu 8988t | C73(1.54); C286(0.98) | LDD1307 | [5] |
| LDCM0429 | CL4 | PaTu 8988t | C73(1.14); C286(0.93) | LDD1308 | [5] |
| LDCM0430 | CL40 | PaTu 8988t | C73(1.23); C286(1.04) | LDD1309 | [5] |
| LDCM0431 | CL41 | PaTu 8988t | C73(1.73); C286(1.14) | LDD1310 | [5] |
| LDCM0432 | CL42 | PaTu 8988t | C73(1.74); C286(1.30) | LDD1311 | [5] |
| LDCM0433 | CL43 | PaTu 8988t | C73(2.27); C286(1.38) | LDD1312 | [5] |
| LDCM0434 | CL44 | PaTu 8988t | C73(1.78); C286(1.11) | LDD1313 | [5] |
| LDCM0435 | CL45 | PaTu 8988t | C73(1.10); C286(1.03) | LDD1314 | [5] |
| LDCM0436 | CL46 | PaTu 8988t | C73(1.06); C286(1.01) | LDD1315 | [5] |
| LDCM0437 | CL47 | PaTu 8988t | C73(0.93); C286(1.03) | LDD1316 | [5] |
| LDCM0438 | CL48 | PaTu 8988t | C73(0.95); C286(1.05) | LDD1317 | [5] |
| LDCM0439 | CL49 | PaTu 8988t | C73(1.07); C286(1.00) | LDD1318 | [5] |
| LDCM0440 | CL5 | PaTu 8988t | C73(1.63); C286(0.89) | LDD1319 | [5] |
| LDCM0441 | CL50 | PaTu 8988t | C73(1.08); C286(0.91) | LDD1320 | [5] |
| LDCM0442 | CL51 | PaTu 8988t | C73(0.99); C286(0.98) | LDD1321 | [5] |
| LDCM0443 | CL52 | PaTu 8988t | C73(1.03); C286(1.05) | LDD1322 | [5] |
| LDCM0444 | CL53 | PaTu 8988t | C73(1.22); C286(1.01) | LDD1323 | [5] |
| LDCM0445 | CL54 | PaTu 8988t | C73(0.85); C286(1.09) | LDD1324 | [5] |
| LDCM0446 | CL55 | PaTu 8988t | C73(1.00); C286(0.99) | LDD1325 | [5] |
| LDCM0447 | CL56 | PaTu 8988t | C73(0.76); C286(1.07) | LDD1326 | [5] |
| LDCM0448 | CL57 | PaTu 8988t | C73(1.02); C286(0.96) | LDD1327 | [5] |
| LDCM0449 | CL58 | PaTu 8988t | C73(0.97); C286(0.95) | LDD1328 | [5] |
| LDCM0450 | CL59 | PaTu 8988t | C73(0.99); C286(0.94) | LDD1329 | [5] |
| LDCM0451 | CL6 | PaTu 8988t | C73(1.23); C286(0.89) | LDD1330 | [5] |
| LDCM0452 | CL60 | PaTu 8988t | C73(0.94); C286(1.03) | LDD1331 | [5] |
| LDCM0453 | CL61 | HCT 116 | C286(0.90) | LDD0770 | [5] |
| LDCM0454 | CL62 | HCT 116 | C286(0.88) | LDD0771 | [5] |
| LDCM0455 | CL63 | HCT 116 | C286(1.01) | LDD0772 | [5] |
| LDCM0456 | CL64 | HCT 116 | C286(1.01) | LDD0773 | [5] |
| LDCM0457 | CL65 | HCT 116 | C286(0.94) | LDD0774 | [5] |
| LDCM0458 | CL66 | HCT 116 | C286(1.03) | LDD0775 | [5] |
| LDCM0459 | CL67 | HCT 116 | C286(1.00) | LDD0776 | [5] |
| LDCM0460 | CL68 | HCT 116 | C286(0.93) | LDD0777 | [5] |
| LDCM0461 | CL69 | HCT 116 | C286(1.02) | LDD0778 | [5] |
| LDCM0462 | CL7 | PaTu 8988t | C73(1.31); C286(0.99) | LDD1341 | [5] |
| LDCM0463 | CL70 | HCT 116 | C286(0.91) | LDD0780 | [5] |
| LDCM0464 | CL71 | HCT 116 | C286(1.06) | LDD0781 | [5] |
| LDCM0465 | CL72 | HCT 116 | C286(1.02) | LDD0782 | [5] |
| LDCM0466 | CL73 | HCT 116 | C286(0.81) | LDD0783 | [5] |
| LDCM0467 | CL74 | HCT 116 | C286(0.86) | LDD0784 | [5] |
| LDCM0469 | CL76 | PaTu 8988t | C286(1.17) | LDD1348 | [5] |
| LDCM0470 | CL77 | PaTu 8988t | C286(0.71) | LDD1349 | [5] |
| LDCM0471 | CL78 | PaTu 8988t | C286(0.83) | LDD1350 | [5] |
| LDCM0472 | CL79 | PaTu 8988t | C286(0.92) | LDD1351 | [5] |
| LDCM0473 | CL8 | PaTu 8988t | C73(1.34); C286(0.82) | LDD1352 | [5] |
| LDCM0474 | CL80 | PaTu 8988t | C286(1.02) | LDD1353 | [5] |
| LDCM0475 | CL81 | PaTu 8988t | C286(1.04) | LDD1354 | [5] |
| LDCM0476 | CL82 | PaTu 8988t | C286(0.82) | LDD1355 | [5] |
| LDCM0477 | CL83 | PaTu 8988t | C286(0.98) | LDD1356 | [5] |
| LDCM0478 | CL84 | PaTu 8988t | C286(1.13) | LDD1357 | [5] |
| LDCM0479 | CL85 | PaTu 8988t | C286(0.90) | LDD1358 | [5] |
| LDCM0480 | CL86 | PaTu 8988t | C286(0.87) | LDD1359 | [5] |
| LDCM0481 | CL87 | PaTu 8988t | C286(0.98) | LDD1360 | [5] |
| LDCM0482 | CL88 | PaTu 8988t | C286(0.92) | LDD1361 | [5] |
| LDCM0483 | CL89 | PaTu 8988t | C286(0.93) | LDD1362 | [5] |
| LDCM0484 | CL9 | PaTu 8988t | C73(1.24); C286(0.85) | LDD1363 | [5] |
| LDCM0485 | CL90 | PaTu 8988t | C286(1.02) | LDD1364 | [5] |
| LDCM0486 | CL91 | HEK-293T | C286(0.91); C73(1.02); C245(0.97) | LDD1689 | [16] |
| LDCM0487 | CL92 | HEK-293T | C286(0.98); C73(0.97); C245(0.96) | LDD1690 | [16] |
| LDCM0488 | CL93 | HEK-293T | C286(0.99); C73(1.08); C245(1.26) | LDD1691 | [16] |
| LDCM0489 | CL94 | HEK-293T | C286(0.99); C73(1.06) | LDD1692 | [16] |
| LDCM0490 | CL95 | HEK-293T | C286(1.10); C73(0.94); C245(1.35) | LDD1693 | [16] |
| LDCM0491 | CL96 | HEK-293T | C286(1.07); C73(0.99); C245(0.98) | LDD1694 | [16] |
| LDCM0492 | CL97 | HEK-293T | C286(0.98); C73(0.90); C245(1.04) | LDD1695 | [16] |
| LDCM0493 | CL98 | HEK-293T | C286(1.01); C73(0.99); C245(0.93) | LDD1696 | [16] |
| LDCM0494 | CL99 | HEK-293T | C286(0.97); C73(0.98); C245(0.95) | LDD1697 | [16] |
| LDCM0495 | E2913 | HEK-293T | C286(0.92); C73(1.00); C245(1.16) | LDD1698 | [16] |
| LDCM0468 | Fragment33 | HCT 116 | C286(0.87) | LDD0785 | [5] |
| LDCM0427 | Fragment51 | PaTu 8988t | C73(2.27); C286(0.94) | LDD1306 | [5] |
| LDCM0022 | KB02 | HCT 116 | C73(1.07) | LDD0080 | [5] |
| LDCM0023 | KB03 | HCT 116 | C73(1.69) | LDD0081 | [5] |
| LDCM0024 | KB05 | HCT 116 | C73(1.65) | LDD0082 | [5] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C286(0.84) | LDD2093 | [3] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C286(0.32) | LDD2109 | [3] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C286(1.03) | LDD2111 | [3] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C286(0.79) | LDD2127 | [3] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C286(0.94) | LDD2136 | [3] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C286(0.64) | LDD2140 | [3] |
References
