Details of the Target
General Information of Target
| Target ID | LDTP12252 | |||||
|---|---|---|---|---|---|---|
| Target Name | Chromodomain-helicase-DNA-binding protein 8 (CHD8) | |||||
| Gene Name | CHD8 | |||||
| Gene ID | 57680 | |||||
| Synonyms |
HELSNF1; KIAA1564; Chromodomain-helicase-DNA-binding protein 8; CHD-8; EC 3.6.4.12; ATP-dependent helicase CHD8; Helicase with SNF2 domain 1 |
|||||
| 3D Structure | ||||||
| Sequence |
MEALGPGPPASLFQPPRRPGLGTVGKPIRLLANHFQVQIPKIDVYHYDVDIKPEKRPRRV
NREVVDTMVRHFKMQIFGDRQPGYDGKRNMYTAHPLPIGRDRVDMEVTLPGEGKDQTFKV SVQWVSVVSLQLLLEALAGHLNEVPDDSVQALDVITRHLPSMRYTPVGRSFFSPPEGYYH PLGGGREVWFGFHQSVRPAMWNMMLNIDVSATAFYRAQPIIEFMCEVLDIQNINEQTKPL TDSQRVKFTKEIRGLKVEVTHCGQMKRKYRVCNVTRRPASHQTFPLQLENGQAMECTVAQ YFKQKYSLQLKYPHLPCLQVGQEQKHTYLPLEVCNIVAGQRCIKKLTDNQTSTMIKATAR SAPDRQEEISRLVKSNSMVGGPDPYLKEFGIVVHNEMTELTGRVLPAPMLQYGGRNKTVA TPNQGVWDMRGKQFYAGIEIKVWAVACFAPQKQCREDLLKSFTDQLRKISKDAGMPIQGQ PCFCKYAQGADSVEPMFKHLKMTYVGLQLIVVILPGKTPVYAEVKRVGDTLLGMATQCVQ VKNVVKTSPQTLSNLCLKINAKLGGINNVLVPHQRPSVFQQPVIFLGADVTHPPAGDGKK PSIAAVVGSMDGHPSRYCATVRVQTSRQEISQELLYSQEVIQDLTNMVRELLIQFYKSTR FKPTRIIYYRGGVSEGQMKQVAWPELIAIRKACISLEEDYRPGITYIVVQKRHHTRLFCA DKTERVGKSGNVPAGTTVDSTITHPSEFDFYLCSHAGIQGTSRPSHYQVLWDDNCFTADE LQLLTYQLCHTYVRCTRSVSIPAPAYYARLVAFRARYHLVDKDHDSAEGSHVSGQSNGRD PQALAKAVQIHHDTQHTMYFA |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
SNF2/RAD54 helicase family, CHD8 subfamily
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
DNA helicase that acts as a chromatin remodeling factor and regulates transcription. Acts as a transcription repressor by remodeling chromatin structure and recruiting histone H1 to target genes. Suppresses p53/TP53-mediated apoptosis by recruiting histone H1 and preventing p53/TP53 transactivation activity. Acts as a negative regulator of Wnt signaling pathway by regulating beta-catenin (CTNNB1) activity. Negatively regulates CTNNB1-targeted gene expression by being recruited specifically to the promoter regions of several CTNNB1 responsive genes. Involved in both enhancer blocking and epigenetic remodeling at chromatin boundary via its interaction with CTCF. Acts as a suppressor of STAT3 activity by suppressing the LIF-induced STAT3 transcriptional activity. Also acts as a transcription activator via its interaction with ZNF143 by participating in efficient U6 RNA polymerase III transcription. Regulates alternative splicing of a core group of genes involved in neuronal differentiation, cell cycle and DNA repair. Enables H3K36me3-coupled transcription elongation and co-transcriptional RNA processing likely via interaction with HNRNPL. {|HAMAP-Rule:MF_03071}.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| CAL51 | Deletion: p.R2352DfsTer3 | DBIA Probe Info | |||
| CCK81 | Deletion: p.K443RfsTer23 | . | |||
| EFO27 | Deletion: p.K2025_L2026del | DBIA Probe Info | |||
| HCT116 | Deletion: p.I351SfsTer23;p.L1905FfsTer30 SNV: p.G1075W;p.R2243C |
DBIA Probe Info | |||
| HCT15 | SNV: p.L190M | . | |||
| HEC1B | SNV: p.R286H; p.R2163H | . | |||
| HT115 | SNV: p.R463W; p.F668S | . | |||
| IGROV1 | SNV: p.E597D | DBIA Probe Info | |||
| Ishikawa (Heraklio) 02 ER | SNV: p.F988C | DBIA Probe Info | |||
| JURKAT | SNV: p.V2045L | . | |||
| JVM3 | SNV: p.A470T | DBIA Probe Info | |||
| L428 | SNV: p.Q1700Ter | DBIA Probe Info | |||
| LNCaP clone FGC | SNV: p.D1426G | DBIA Probe Info | |||
| MDAMB453 | SNV: p.E2027K | . | |||
| MELJUSO | SNV: p.P2345L | DBIA Probe Info | |||
| MEWO | SNV: p.R546C | DBIA Probe Info | |||
| MFE319 | SNV: p.Y675C; p.T976I; p.A1127V | . | |||
| NCIH1299 | SNV: p.R1337Q | . | |||
| NCIH2172 | SNV: p.D1139G | . | |||
| OVCAR5 | SNV: p.R1443C | DBIA Probe Info | |||
| TE11 | Deletion: p.S389_Q392del | DBIA Probe Info | |||
| TYKNU | SNV: p.W1798S | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
BTD Probe Info |
 |
C1780(1.09) | LDD2092 | [1] | |
|
AHL-Pu-1 Probe Info |
 |
C2378(3.06); C1780(2.59) | LDD0168 | [2] | |
|
DBIA Probe Info |
 |
C1116(1.41) | LDD0080 | [3] | |
|
CY4 Probe Info |
 |
N.A. | LDD0247 | [4] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [5] | |
|
Lodoacetamide azide Probe Info |
 |
C1161(0.00); C1947(0.00) | LDD0037 | [6] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [7] | |
|
NPM Probe Info |
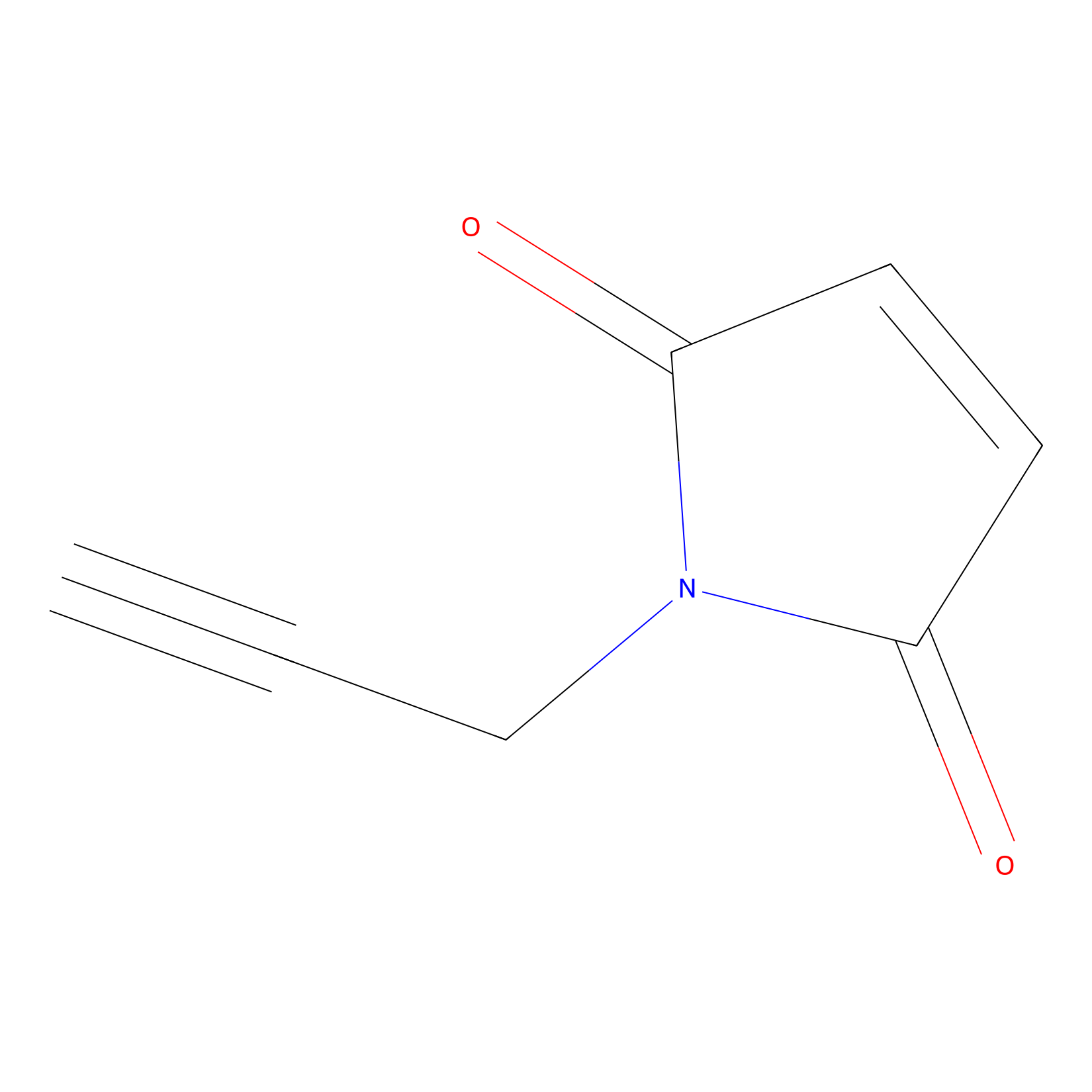 |
N.A. | LDD0016 | [7] | |
|
TFBX Probe Info |
 |
C1980(0.00); C1655(0.00); C1780(0.00); C1116(0.00) | LDD0148 | [8] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [7] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [9] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [9] | |
|
AOyne Probe Info |
 |
12.40 | LDD0443 | [10] | |
|
NAIA_5 Probe Info |
 |
C1980(0.00); C1902(0.00) | LDD2223 | [11] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C1780(0.78) | LDD2142 | [1] |
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C1780(1.03) | LDD2112 | [1] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C1780(0.67) | LDD2117 | [1] |
| LDCM0025 | 4SU-RNA | HEK-293T | C2378(3.06); C1780(2.59) | LDD0168 | [2] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C2378(2.48); C1780(2.54) | LDD0169 | [2] |
| LDCM0214 | AC1 | HCT 116 | C906(1.19) | LDD0531 | [3] |
| LDCM0215 | AC10 | HCT 116 | C906(0.99) | LDD0532 | [3] |
| LDCM0216 | AC100 | HEK-293T | C906(1.00) | LDD0814 | [3] |
| LDCM0217 | AC101 | HEK-293T | C906(1.02) | LDD0815 | [3] |
| LDCM0218 | AC102 | HEK-293T | C906(0.92) | LDD0816 | [3] |
| LDCM0219 | AC103 | HEK-293T | C906(1.11) | LDD0817 | [3] |
| LDCM0220 | AC104 | HEK-293T | C906(0.99) | LDD0818 | [3] |
| LDCM0221 | AC105 | HEK-293T | C906(1.06) | LDD0819 | [3] |
| LDCM0222 | AC106 | HEK-293T | C906(1.01) | LDD0820 | [3] |
| LDCM0223 | AC107 | HEK-293T | C906(0.87) | LDD0821 | [3] |
| LDCM0224 | AC108 | HEK-293T | C906(1.03) | LDD0822 | [3] |
| LDCM0225 | AC109 | HEK-293T | C906(1.01) | LDD0823 | [3] |
| LDCM0226 | AC11 | HCT 116 | C906(0.95) | LDD0543 | [3] |
| LDCM0227 | AC110 | HEK-293T | C906(0.97) | LDD0825 | [3] |
| LDCM0228 | AC111 | HEK-293T | C906(0.93) | LDD0826 | [3] |
| LDCM0229 | AC112 | HEK-293T | C906(1.02) | LDD0827 | [3] |
| LDCM0230 | AC113 | HCT 116 | C906(1.10) | LDD0547 | [3] |
| LDCM0231 | AC114 | HCT 116 | C906(1.10) | LDD0548 | [3] |
| LDCM0232 | AC115 | HCT 116 | C906(0.99) | LDD0549 | [3] |
| LDCM0233 | AC116 | HCT 116 | C906(1.03) | LDD0550 | [3] |
| LDCM0234 | AC117 | HCT 116 | C906(0.95) | LDD0551 | [3] |
| LDCM0235 | AC118 | HCT 116 | C906(1.04) | LDD0552 | [3] |
| LDCM0236 | AC119 | HCT 116 | C906(0.95) | LDD0553 | [3] |
| LDCM0237 | AC12 | HCT 116 | C906(0.97) | LDD0554 | [3] |
| LDCM0238 | AC120 | HCT 116 | C906(1.12) | LDD0555 | [3] |
| LDCM0239 | AC121 | HCT 116 | C906(1.12) | LDD0556 | [3] |
| LDCM0240 | AC122 | HCT 116 | C906(0.87) | LDD0557 | [3] |
| LDCM0241 | AC123 | HCT 116 | C906(1.32) | LDD0558 | [3] |
| LDCM0242 | AC124 | HCT 116 | C906(1.27) | LDD0559 | [3] |
| LDCM0243 | AC125 | HCT 116 | C906(1.10) | LDD0560 | [3] |
| LDCM0244 | AC126 | HCT 116 | C906(1.27) | LDD0561 | [3] |
| LDCM0245 | AC127 | HCT 116 | C906(1.18) | LDD0562 | [3] |
| LDCM0246 | AC128 | HCT 116 | C906(1.27) | LDD0563 | [3] |
| LDCM0247 | AC129 | HCT 116 | C906(1.36) | LDD0564 | [3] |
| LDCM0249 | AC130 | HCT 116 | C906(1.08) | LDD0566 | [3] |
| LDCM0250 | AC131 | HCT 116 | C906(1.32) | LDD0567 | [3] |
| LDCM0251 | AC132 | HCT 116 | C906(1.24) | LDD0568 | [3] |
| LDCM0252 | AC133 | HCT 116 | C906(1.27) | LDD0569 | [3] |
| LDCM0253 | AC134 | HCT 116 | C906(1.24) | LDD0570 | [3] |
| LDCM0254 | AC135 | HCT 116 | C906(1.37) | LDD0571 | [3] |
| LDCM0255 | AC136 | HCT 116 | C906(1.11) | LDD0572 | [3] |
| LDCM0256 | AC137 | HCT 116 | C906(1.21) | LDD0573 | [3] |
| LDCM0257 | AC138 | HCT 116 | C906(1.35) | LDD0574 | [3] |
| LDCM0258 | AC139 | HCT 116 | C906(1.24) | LDD0575 | [3] |
| LDCM0259 | AC14 | HCT 116 | C906(1.10) | LDD0576 | [3] |
| LDCM0260 | AC140 | HCT 116 | C906(1.39) | LDD0577 | [3] |
| LDCM0261 | AC141 | HCT 116 | C906(1.34) | LDD0578 | [3] |
| LDCM0262 | AC142 | HCT 116 | C906(1.17) | LDD0579 | [3] |
| LDCM0263 | AC143 | HCT 116 | C906(0.93) | LDD0580 | [3] |
| LDCM0264 | AC144 | HCT 116 | C906(0.86) | LDD0581 | [3] |
| LDCM0265 | AC145 | HCT 116 | C906(0.96) | LDD0582 | [3] |
| LDCM0266 | AC146 | HCT 116 | C906(1.10) | LDD0583 | [3] |
| LDCM0267 | AC147 | HCT 116 | C906(0.70) | LDD0584 | [3] |
| LDCM0268 | AC148 | HCT 116 | C906(1.78) | LDD0585 | [3] |
| LDCM0269 | AC149 | HCT 116 | C906(1.59) | LDD0586 | [3] |
| LDCM0270 | AC15 | HCT 116 | C906(0.99) | LDD0587 | [3] |
| LDCM0271 | AC150 | HCT 116 | C906(0.88) | LDD0588 | [3] |
| LDCM0272 | AC151 | HCT 116 | C906(0.67) | LDD0589 | [3] |
| LDCM0273 | AC152 | HCT 116 | C906(0.81) | LDD0590 | [3] |
| LDCM0274 | AC153 | HCT 116 | C906(1.21) | LDD0591 | [3] |
| LDCM0621 | AC154 | HCT 116 | C906(0.96) | LDD2158 | [3] |
| LDCM0622 | AC155 | HCT 116 | C906(1.06) | LDD2159 | [3] |
| LDCM0623 | AC156 | HCT 116 | C906(0.90) | LDD2160 | [3] |
| LDCM0624 | AC157 | HCT 116 | C906(0.87) | LDD2161 | [3] |
| LDCM0276 | AC17 | HCT 116 | C906(0.93) | LDD0593 | [3] |
| LDCM0277 | AC18 | HCT 116 | C906(1.21) | LDD0594 | [3] |
| LDCM0278 | AC19 | HCT 116 | C906(0.95) | LDD0595 | [3] |
| LDCM0279 | AC2 | HCT 116 | C906(1.28) | LDD0596 | [3] |
| LDCM0280 | AC20 | HCT 116 | C906(1.01) | LDD0597 | [3] |
| LDCM0281 | AC21 | HCT 116 | C906(1.12) | LDD0598 | [3] |
| LDCM0282 | AC22 | HCT 116 | C906(0.94) | LDD0599 | [3] |
| LDCM0283 | AC23 | HCT 116 | C906(0.94) | LDD0600 | [3] |
| LDCM0284 | AC24 | HCT 116 | C906(0.90) | LDD0601 | [3] |
| LDCM0285 | AC25 | HCT 116 | C1116(0.74); C906(0.98) | LDD0602 | [3] |
| LDCM0286 | AC26 | HCT 116 | C1116(0.83); C906(0.90) | LDD0603 | [3] |
| LDCM0287 | AC27 | HCT 116 | C1116(0.84); C906(0.91) | LDD0604 | [3] |
| LDCM0288 | AC28 | HCT 116 | C1116(0.85); C906(0.94) | LDD0605 | [3] |
| LDCM0289 | AC29 | HCT 116 | C906(0.87); C1116(0.99) | LDD0606 | [3] |
| LDCM0290 | AC3 | HCT 116 | C906(1.13) | LDD0607 | [3] |
| LDCM0291 | AC30 | HCT 116 | C906(0.97); C1116(1.14) | LDD0608 | [3] |
| LDCM0292 | AC31 | HCT 116 | C1116(0.83); C906(0.92) | LDD0609 | [3] |
| LDCM0293 | AC32 | HCT 116 | C906(0.97); C1116(1.10) | LDD0610 | [3] |
| LDCM0294 | AC33 | HCT 116 | C1116(0.84); C906(0.88) | LDD0611 | [3] |
| LDCM0295 | AC34 | HCT 116 | C1116(0.82); C906(0.90) | LDD0612 | [3] |
| LDCM0296 | AC35 | HCT 116 | C906(0.91) | LDD0613 | [3] |
| LDCM0297 | AC36 | HCT 116 | C906(0.79) | LDD0614 | [3] |
| LDCM0298 | AC37 | HCT 116 | C906(0.89) | LDD0615 | [3] |
| LDCM0299 | AC38 | HCT 116 | C906(0.95) | LDD0616 | [3] |
| LDCM0300 | AC39 | HCT 116 | C906(0.86) | LDD0617 | [3] |
| LDCM0301 | AC4 | HCT 116 | C906(1.29) | LDD0618 | [3] |
| LDCM0302 | AC40 | HCT 116 | C906(0.92) | LDD0619 | [3] |
| LDCM0303 | AC41 | HCT 116 | C906(0.88) | LDD0620 | [3] |
| LDCM0304 | AC42 | HCT 116 | C906(1.01) | LDD0621 | [3] |
| LDCM0305 | AC43 | HCT 116 | C906(0.99) | LDD0622 | [3] |
| LDCM0306 | AC44 | HCT 116 | C906(0.93) | LDD0623 | [3] |
| LDCM0307 | AC45 | HCT 116 | C906(0.85) | LDD0624 | [3] |
| LDCM0308 | AC46 | HCT 116 | C906(0.86) | LDD0625 | [3] |
| LDCM0309 | AC47 | HCT 116 | C906(0.82) | LDD0626 | [3] |
| LDCM0310 | AC48 | HCT 116 | C906(0.81) | LDD0627 | [3] |
| LDCM0311 | AC49 | HCT 116 | C906(0.91) | LDD0628 | [3] |
| LDCM0312 | AC5 | HCT 116 | C906(1.34) | LDD0629 | [3] |
| LDCM0313 | AC50 | HCT 116 | C906(1.01) | LDD0630 | [3] |
| LDCM0314 | AC51 | HCT 116 | C906(0.86) | LDD0631 | [3] |
| LDCM0315 | AC52 | HCT 116 | C906(0.83) | LDD0632 | [3] |
| LDCM0316 | AC53 | HCT 116 | C906(0.92) | LDD0633 | [3] |
| LDCM0317 | AC54 | HCT 116 | C906(0.94) | LDD0634 | [3] |
| LDCM0318 | AC55 | HCT 116 | C906(0.88) | LDD0635 | [3] |
| LDCM0319 | AC56 | HCT 116 | C906(1.01) | LDD0636 | [3] |
| LDCM0320 | AC57 | HEK-293T | C906(1.17) | LDD0918 | [3] |
| LDCM0321 | AC58 | HEK-293T | C906(1.14) | LDD0919 | [3] |
| LDCM0322 | AC59 | HEK-293T | C906(1.03) | LDD0920 | [3] |
| LDCM0323 | AC6 | HCT 116 | C906(0.98) | LDD0640 | [3] |
| LDCM0324 | AC60 | HEK-293T | C906(1.18) | LDD0922 | [3] |
| LDCM0325 | AC61 | HEK-293T | C906(0.95) | LDD0923 | [3] |
| LDCM0326 | AC62 | HEK-293T | C906(1.07) | LDD0924 | [3] |
| LDCM0327 | AC63 | HEK-293T | C906(1.00) | LDD0925 | [3] |
| LDCM0328 | AC64 | HEK-293T | C906(1.03) | LDD0926 | [3] |
| LDCM0329 | AC65 | HEK-293T | C906(1.10) | LDD0927 | [3] |
| LDCM0330 | AC66 | HEK-293T | C906(0.93) | LDD0928 | [3] |
| LDCM0331 | AC67 | HEK-293T | C906(1.06) | LDD0929 | [3] |
| LDCM0332 | AC68 | HCT 116 | C906(1.11) | LDD0649 | [3] |
| LDCM0333 | AC69 | HCT 116 | C906(1.11) | LDD0650 | [3] |
| LDCM0334 | AC7 | HCT 116 | C906(0.97) | LDD0651 | [3] |
| LDCM0335 | AC70 | HCT 116 | C906(1.00) | LDD0652 | [3] |
| LDCM0336 | AC71 | HCT 116 | C906(1.10) | LDD0653 | [3] |
| LDCM0337 | AC72 | HCT 116 | C906(1.12) | LDD0654 | [3] |
| LDCM0338 | AC73 | HCT 116 | C906(1.12) | LDD0655 | [3] |
| LDCM0339 | AC74 | HCT 116 | C906(1.14) | LDD0656 | [3] |
| LDCM0340 | AC75 | HCT 116 | C906(1.03) | LDD0657 | [3] |
| LDCM0341 | AC76 | HCT 116 | C906(1.17) | LDD0658 | [3] |
| LDCM0342 | AC77 | HCT 116 | C906(0.96) | LDD0659 | [3] |
| LDCM0343 | AC78 | HCT 116 | C906(1.07) | LDD0660 | [3] |
| LDCM0344 | AC79 | HCT 116 | C906(0.95) | LDD0661 | [3] |
| LDCM0345 | AC8 | HCT 116 | C906(0.99) | LDD0662 | [3] |
| LDCM0346 | AC80 | HCT 116 | C906(1.01) | LDD0663 | [3] |
| LDCM0347 | AC81 | HCT 116 | C906(1.21) | LDD0664 | [3] |
| LDCM0348 | AC82 | HCT 116 | C906(1.28) | LDD0665 | [3] |
| LDCM0349 | AC83 | HCT 116 | C906(1.14) | LDD0666 | [3] |
| LDCM0350 | AC84 | HCT 116 | C906(1.05) | LDD0667 | [3] |
| LDCM0351 | AC85 | HCT 116 | C906(0.93) | LDD0668 | [3] |
| LDCM0352 | AC86 | HCT 116 | C906(0.92) | LDD0669 | [3] |
| LDCM0353 | AC87 | HCT 116 | C906(0.89) | LDD0670 | [3] |
| LDCM0354 | AC88 | HCT 116 | C906(0.86) | LDD0671 | [3] |
| LDCM0355 | AC89 | HCT 116 | C906(1.03) | LDD0672 | [3] |
| LDCM0357 | AC90 | HCT 116 | C906(0.79) | LDD0674 | [3] |
| LDCM0358 | AC91 | HCT 116 | C906(1.04) | LDD0675 | [3] |
| LDCM0359 | AC92 | HCT 116 | C906(1.08) | LDD0676 | [3] |
| LDCM0360 | AC93 | HCT 116 | C906(0.99) | LDD0677 | [3] |
| LDCM0361 | AC94 | HCT 116 | C906(0.91) | LDD0678 | [3] |
| LDCM0362 | AC95 | HCT 116 | C906(0.92) | LDD0679 | [3] |
| LDCM0363 | AC96 | HCT 116 | C906(1.03) | LDD0680 | [3] |
| LDCM0364 | AC97 | HCT 116 | C906(1.21) | LDD0681 | [3] |
| LDCM0365 | AC98 | HEK-293T | C906(1.14) | LDD0963 | [3] |
| LDCM0366 | AC99 | HEK-293T | C906(1.01) | LDD0964 | [3] |
| LDCM0248 | AKOS034007472 | HCT 116 | C906(0.98) | LDD0565 | [3] |
| LDCM0356 | AKOS034007680 | HCT 116 | C906(0.98) | LDD0673 | [3] |
| LDCM0275 | AKOS034007705 | HCT 116 | C906(1.01) | LDD0592 | [3] |
| LDCM0367 | CL1 | HEK-293T | C1116(1.48); C1913(1.72); C906(0.79) | LDD0965 | [3] |
| LDCM0368 | CL10 | HEK-293T | C1116(2.18); C1913(1.39); C906(0.73) | LDD0966 | [3] |
| LDCM0369 | CL100 | HCT 116 | C906(1.23) | LDD0686 | [3] |
| LDCM0370 | CL101 | HCT 116 | C906(0.97) | LDD0687 | [3] |
| LDCM0371 | CL102 | HCT 116 | C906(1.04) | LDD0688 | [3] |
| LDCM0372 | CL103 | HCT 116 | C906(1.08) | LDD0689 | [3] |
| LDCM0373 | CL104 | HCT 116 | C906(0.99) | LDD0690 | [3] |
| LDCM0374 | CL105 | HCT 116 | C906(1.37) | LDD0691 | [3] |
| LDCM0375 | CL106 | HCT 116 | C906(1.65) | LDD0692 | [3] |
| LDCM0376 | CL107 | HCT 116 | C906(1.34) | LDD0693 | [3] |
| LDCM0377 | CL108 | HCT 116 | C906(1.03) | LDD0694 | [3] |
| LDCM0378 | CL109 | HCT 116 | C906(1.40) | LDD0695 | [3] |
| LDCM0379 | CL11 | HEK-293T | C1116(2.14); C1913(1.54); C906(0.86) | LDD0977 | [3] |
| LDCM0380 | CL110 | HCT 116 | C906(1.16) | LDD0697 | [3] |
| LDCM0381 | CL111 | HCT 116 | C906(0.95) | LDD0698 | [3] |
| LDCM0382 | CL112 | HCT 116 | C906(0.91); C1116(0.91) | LDD0699 | [3] |
| LDCM0383 | CL113 | HCT 116 | C1116(0.91); C906(0.94) | LDD0700 | [3] |
| LDCM0384 | CL114 | HCT 116 | C906(0.85); C1116(0.94) | LDD0701 | [3] |
| LDCM0385 | CL115 | HCT 116 | C906(0.90); C1116(1.00) | LDD0702 | [3] |
| LDCM0386 | CL116 | HCT 116 | C1116(0.83); C906(0.94) | LDD0703 | [3] |
| LDCM0387 | CL117 | HCT 116 | C906(1.17) | LDD0704 | [3] |
| LDCM0388 | CL118 | HCT 116 | C906(0.94) | LDD0705 | [3] |
| LDCM0389 | CL119 | HCT 116 | C906(0.99) | LDD0706 | [3] |
| LDCM0390 | CL12 | HEK-293T | C1116(1.18); C1913(1.19); C906(1.06) | LDD0988 | [3] |
| LDCM0391 | CL120 | HCT 116 | C906(0.82) | LDD0708 | [3] |
| LDCM0392 | CL121 | HCT 116 | C906(0.87) | LDD0709 | [3] |
| LDCM0393 | CL122 | HCT 116 | C906(1.09) | LDD0710 | [3] |
| LDCM0394 | CL123 | HCT 116 | C906(0.98) | LDD0711 | [3] |
| LDCM0395 | CL124 | HCT 116 | C906(0.85) | LDD0712 | [3] |
| LDCM0396 | CL125 | HEK-293T | C906(0.87) | LDD0994 | [3] |
| LDCM0397 | CL126 | HEK-293T | C906(1.05) | LDD0995 | [3] |
| LDCM0398 | CL127 | HEK-293T | C906(0.92) | LDD0996 | [3] |
| LDCM0399 | CL128 | HEK-293T | C906(0.83) | LDD0997 | [3] |
| LDCM0400 | CL13 | HEK-293T | C1116(1.16); C1913(1.33); C906(0.98) | LDD0998 | [3] |
| LDCM0401 | CL14 | HEK-293T | C1116(1.40); C1913(1.55); C906(0.86) | LDD0999 | [3] |
| LDCM0402 | CL15 | HEK-293T | C1116(1.72); C1913(1.52); C906(0.79) | LDD1000 | [3] |
| LDCM0403 | CL16 | HCT 116 | C906(0.98) | LDD0720 | [3] |
| LDCM0404 | CL17 | HCT 116 | C906(1.07); C1116(1.09) | LDD0721 | [3] |
| LDCM0405 | CL18 | HCT 116 | C906(0.89); C1116(0.99) | LDD0722 | [3] |
| LDCM0406 | CL19 | HCT 116 | C1116(0.75); C906(1.00) | LDD0723 | [3] |
| LDCM0407 | CL2 | HEK-293T | C1116(1.33); C1913(1.79); C906(0.72) | LDD1005 | [3] |
| LDCM0408 | CL20 | HCT 116 | C906(0.98); C1116(1.10) | LDD0725 | [3] |
| LDCM0409 | CL21 | HCT 116 | C1116(0.83); C906(1.03) | LDD0726 | [3] |
| LDCM0410 | CL22 | HCT 116 | C1116(0.78); C906(1.12) | LDD0727 | [3] |
| LDCM0411 | CL23 | HCT 116 | C906(1.02); C1116(1.26) | LDD0728 | [3] |
| LDCM0412 | CL24 | HCT 116 | C1116(0.86); C906(1.02) | LDD0729 | [3] |
| LDCM0413 | CL25 | HCT 116 | C1116(0.75); C906(1.02) | LDD0730 | [3] |
| LDCM0414 | CL26 | HCT 116 | C1116(0.96); C906(0.97) | LDD0731 | [3] |
| LDCM0415 | CL27 | HCT 116 | C1116(1.21); C906(0.91) | LDD0732 | [3] |
| LDCM0416 | CL28 | HCT 116 | C1116(0.82); C906(0.97) | LDD0733 | [3] |
| LDCM0417 | CL29 | HCT 116 | C1116(0.81); C906(0.99) | LDD0734 | [3] |
| LDCM0418 | CL3 | HEK-293T | C1116(1.30); C1913(1.09); C906(1.24) | LDD1016 | [3] |
| LDCM0419 | CL30 | HCT 116 | C1116(0.94); C906(1.00) | LDD0736 | [3] |
| LDCM0420 | CL31 | HCT 116 | C1116(0.84) | LDD0737 | [3] |
| LDCM0421 | CL32 | HEK-293T | C1913(0.81); C906(1.13) | LDD1019 | [3] |
| LDCM0422 | CL33 | HEK-293T | C1913(1.35); C906(1.25) | LDD1020 | [3] |
| LDCM0423 | CL34 | HEK-293T | C1913(1.40); C906(1.19) | LDD1021 | [3] |
| LDCM0424 | CL35 | HEK-293T | C1913(0.92); C906(1.22) | LDD1022 | [3] |
| LDCM0425 | CL36 | HEK-293T | C1913(0.99); C906(1.08) | LDD1023 | [3] |
| LDCM0426 | CL37 | HEK-293T | C1913(1.04); C906(1.15) | LDD1024 | [3] |
| LDCM0428 | CL39 | HEK-293T | C1913(1.32); C906(1.56) | LDD1026 | [3] |
| LDCM0429 | CL4 | HEK-293T | C1116(1.37); C1913(1.24); C906(0.57) | LDD1027 | [3] |
| LDCM0430 | CL40 | HEK-293T | C1913(1.21); C906(1.18) | LDD1028 | [3] |
| LDCM0431 | CL41 | HEK-293T | C1913(1.16); C906(1.33) | LDD1029 | [3] |
| LDCM0432 | CL42 | HEK-293T | C1913(0.84); C906(1.35) | LDD1030 | [3] |
| LDCM0433 | CL43 | HEK-293T | C1913(1.12); C906(1.40) | LDD1031 | [3] |
| LDCM0434 | CL44 | HEK-293T | C1913(0.95); C906(1.09) | LDD1032 | [3] |
| LDCM0435 | CL45 | HEK-293T | C1913(1.37); C906(1.37) | LDD1033 | [3] |
| LDCM0436 | CL46 | HCT 116 | C906(0.76) | LDD0753 | [3] |
| LDCM0437 | CL47 | HCT 116 | C906(0.77) | LDD0754 | [3] |
| LDCM0438 | CL48 | HCT 116 | C906(0.74) | LDD0755 | [3] |
| LDCM0439 | CL49 | HCT 116 | C906(0.84) | LDD0756 | [3] |
| LDCM0440 | CL5 | HEK-293T | C1116(1.34); C1913(1.00); C906(0.78) | LDD1038 | [3] |
| LDCM0441 | CL50 | HCT 116 | C906(0.82) | LDD0758 | [3] |
| LDCM0442 | CL51 | HCT 116 | C906(0.80) | LDD0759 | [3] |
| LDCM0443 | CL52 | HCT 116 | C906(0.76) | LDD0760 | [3] |
| LDCM0444 | CL53 | HCT 116 | C906(0.79) | LDD0761 | [3] |
| LDCM0445 | CL54 | HCT 116 | C906(0.72) | LDD0762 | [3] |
| LDCM0446 | CL55 | HCT 116 | C906(0.72) | LDD0763 | [3] |
| LDCM0447 | CL56 | HCT 116 | C906(0.90) | LDD0764 | [3] |
| LDCM0448 | CL57 | HCT 116 | C906(0.76) | LDD0765 | [3] |
| LDCM0449 | CL58 | HCT 116 | C906(0.77) | LDD0766 | [3] |
| LDCM0450 | CL59 | HCT 116 | C906(0.95) | LDD0767 | [3] |
| LDCM0451 | CL6 | HEK-293T | C1116(1.42); C1913(1.72); C906(0.80) | LDD1049 | [3] |
| LDCM0452 | CL60 | HCT 116 | C906(0.88) | LDD0769 | [3] |
| LDCM0453 | CL61 | HCT 116 | C1116(0.92); C906(1.29) | LDD0770 | [3] |
| LDCM0454 | CL62 | HCT 116 | C1116(0.86); C906(1.25) | LDD0771 | [3] |
| LDCM0455 | CL63 | HCT 116 | C1116(0.83); C906(1.10) | LDD0772 | [3] |
| LDCM0456 | CL64 | HCT 116 | C1116(0.86); C906(1.12) | LDD0773 | [3] |
| LDCM0457 | CL65 | HCT 116 | C1116(0.89); C906(1.12) | LDD0774 | [3] |
| LDCM0458 | CL66 | HCT 116 | C1116(0.97); C906(1.50) | LDD0775 | [3] |
| LDCM0459 | CL67 | HCT 116 | C1116(0.96); C906(1.41) | LDD0776 | [3] |
| LDCM0460 | CL68 | HCT 116 | C1116(1.12); C906(1.43) | LDD0777 | [3] |
| LDCM0461 | CL69 | HCT 116 | C1116(0.83); C906(1.47) | LDD0778 | [3] |
| LDCM0462 | CL7 | HEK-293T | C1116(1.43); C1913(1.10); C906(1.24) | LDD1060 | [3] |
| LDCM0463 | CL70 | HCT 116 | C1116(0.94); C906(1.35) | LDD0780 | [3] |
| LDCM0464 | CL71 | HCT 116 | C1116(0.89); C906(1.39) | LDD0781 | [3] |
| LDCM0465 | CL72 | HCT 116 | C1116(0.97); C906(1.08) | LDD0782 | [3] |
| LDCM0466 | CL73 | HCT 116 | C1116(0.94); C906(1.36) | LDD0783 | [3] |
| LDCM0467 | CL74 | HCT 116 | C1116(0.91); C906(1.25) | LDD0784 | [3] |
| LDCM0469 | CL76 | HCT 116 | C1116(0.90) | LDD0786 | [3] |
| LDCM0470 | CL77 | HCT 116 | C1116(1.00) | LDD0787 | [3] |
| LDCM0471 | CL78 | HCT 116 | C1116(0.97) | LDD0788 | [3] |
| LDCM0472 | CL79 | HCT 116 | C1116(1.13) | LDD0789 | [3] |
| LDCM0473 | CL8 | HEK-293T | C1116(1.60); C906(0.87) | LDD1071 | [3] |
| LDCM0474 | CL80 | HCT 116 | C1116(1.00) | LDD0791 | [3] |
| LDCM0475 | CL81 | HCT 116 | C1116(0.87) | LDD0792 | [3] |
| LDCM0476 | CL82 | HCT 116 | C1116(0.94) | LDD0793 | [3] |
| LDCM0477 | CL83 | HCT 116 | C1116(0.93) | LDD0794 | [3] |
| LDCM0478 | CL84 | HCT 116 | C1116(0.97) | LDD0795 | [3] |
| LDCM0479 | CL85 | HCT 116 | C1116(1.09) | LDD0796 | [3] |
| LDCM0480 | CL86 | HCT 116 | C1116(0.94) | LDD0797 | [3] |
| LDCM0481 | CL87 | HCT 116 | C1116(0.96) | LDD0798 | [3] |
| LDCM0482 | CL88 | HCT 116 | C1116(0.92) | LDD0799 | [3] |
| LDCM0483 | CL89 | HCT 116 | C1116(0.92) | LDD0800 | [3] |
| LDCM0484 | CL9 | HEK-293T | C1116(1.32); C906(0.81) | LDD1082 | [3] |
| LDCM0485 | CL90 | HCT 116 | C1116(1.13) | LDD0802 | [3] |
| LDCM0486 | CL91 | HCT 116 | C906(1.17) | LDD0803 | [3] |
| LDCM0487 | CL92 | HCT 116 | C906(1.26) | LDD0804 | [3] |
| LDCM0488 | CL93 | HCT 116 | C906(1.06) | LDD0805 | [3] |
| LDCM0489 | CL94 | HCT 116 | C906(1.44) | LDD0806 | [3] |
| LDCM0490 | CL95 | HCT 116 | C906(1.46) | LDD0807 | [3] |
| LDCM0491 | CL96 | HCT 116 | C906(1.21) | LDD0808 | [3] |
| LDCM0492 | CL97 | HCT 116 | C906(1.23) | LDD0809 | [3] |
| LDCM0493 | CL98 | HCT 116 | C906(1.13) | LDD0810 | [3] |
| LDCM0494 | CL99 | HCT 116 | C906(1.34) | LDD0811 | [3] |
| LDCM0495 | E2913 | HEK-293T | C1691(0.99); C1913(0.90); C2300(0.94); C1780(1.02) | LDD1698 | [12] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C1780(0.74) | LDD1702 | [1] |
| LDCM0468 | Fragment33 | HCT 116 | C1116(0.85); C906(1.15) | LDD0785 | [3] |
| LDCM0427 | Fragment51 | HEK-293T | C1913(0.94); C906(1.09) | LDD1025 | [3] |
| LDCM0022 | KB02 | HCT 116 | C1116(1.41) | LDD0080 | [3] |
| LDCM0023 | KB03 | HCT 116 | C1116(1.63) | LDD0081 | [3] |
| LDCM0024 | KB05 | HCT 116 | C1116(2.67) | LDD0082 | [3] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C1780(1.09) | LDD2092 | [1] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C1780(1.41) | LDD2098 | [1] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C1780(1.24) | LDD2099 | [1] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C1780(0.74) | LDD2104 | [1] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C1780(1.17) | LDD2105 | [1] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C1780(0.85) | LDD2108 | [1] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C1780(0.66) | LDD2114 | [1] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C1780(0.99) | LDD2123 | [1] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C1780(1.66) | LDD2136 | [1] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C1780(1.11) | LDD2140 | [1] |
| LDCM0551 | Nucleophilic fragment 5b | MDA-MB-231 | C1780(4.11) | LDD2145 | [1] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Zinc finger and BTB domain-containing protein 7A (ZBTB7A) | . | O95365 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| WD repeat-containing protein 5 (WDR5) | WD repeat WDR5/wds family | P61964 | |||
| Retinoblastoma-binding protein 5 (RBBP5) | . | Q15291 | |||
References
