Details of the Target
General Information of Target
| Target ID | LDTP09702 | |||||
|---|---|---|---|---|---|---|
| Target Name | Nesprin-2 (SYNE2) | |||||
| Gene Name | SYNE2 | |||||
| Gene ID | 23224 | |||||
| Synonyms |
KIAA1011; NUA; Nesprin-2; KASH domain-containing protein 2; KASH2; Nuclear envelope spectrin repeat protein 2; Nucleus and actin connecting element protein; Protein NUANCE; Synaptic nuclear envelope protein 2; Syne-2
|
|||||
| 3D Structure | ||||||
| Sequence |
MASSPELPTEDEQGSWGIDDLHISLQAEQEDTQKKAFTCWINSQLARHTSPSVISDLFTD
IKKGHVLLDLLEVLSGQQLPRDKGSNTFQCRINIEHALTFLRNRSIKLINIHVTDIIDGN PSIILGLIWTIILHFHIEKLAQTLSCNYNQPSLDDVSVVDSSPASSPPAKKCSKVQARWQ MSARKALLLWAQEQCATYESVNVTDFKSSWRNGMAFLAIIHALRPDLIDMKSVKHRSNKD NLREAFRIAEQELKIPRLLEPEDVDVVDPDEKSIMTYVAQFLQYSKDAPGTGEEAQGKVK DAMGWLTLQKEKLQKLLKDSENDTYFKKYNSLLSFMESFNEEKKSFLDVLSIKRDLDELD KDHLQLREAWDGLDHQINAWKIKLNYALPPPLHQTEAWLQEVEELMDEDLSASQDHSQAV TLIQEKMTLFKSLMDRFEHHSNILLTFENKDENHLPLVPPNKLEEMKRRINNILEKKFIL LLEFHYYKCLVLGLVDEVKSKLDIWNIKYGSRESVELLLEDWHKFIEEKEFLARLDTSFQ KCGEIYKNLAGECQNINKQYMMVKSDVCMYRKNIYNVKSTLQKVLACWATYVENLRLLRA CFEETKKEEIKEVPFETLAQWNLEHATLNEAGNFLVEVSNDVVGSSISKELRRLNKRWRK LVSKTQLEMNLPLMIKKQDQPTFDNSGNILSKEEKATVEFSTDMSVELPENYNQNIKAGE KHEKENEEFTGQLKVAKDVEKLIGQVEIWEAEAKSVLDQDDVDTSMEESLKHLIAKGSMF DELMARSEDMLQMDIQNISSQESFQHVLTTGLQAKIQEAKEKVQINVVKLIAALKNLTDV SPDLDIRLKMEESQKELESYMMRAQQLLGQRESPGELISKHKEALIISNTKSLAKYLKAV EELKNNVTEDIKMSLEEKSRDVCAKWESLHHELSLYVQQLKIDIEKGKLSDNILKLEKQI NKEKKLIRRGRTKGLIKEHEACFSEEGCLYQLNHHMEVLRELCEELPSQKSQQEVKRLLK DYEQKIERLLKCASEIHMTLQPTAGGTSKNEGTITTSENRGGDPHSEAPFAKSDNQPSTE KAMEPTMKFSLASVLRPLQEESIMEKDYSASINSLLERYDTYRDILEHHLQNNKFRITSD FSSEEDRSSSCLQAKLTDLQVIKNETDARWKEFEIISLKLENHVNDIKKPFVIKERDTLK ERERELQMTLNTRMESLETALRLVLPVEKASLLLCGSDLPLHKMAIQGFHLIDADRIYQH LRNIQDSIAKQIEICNRLEEPGNFVLKELHPFDLHAMQNIILKYKTQFEGMNHRVQRSED TLKALEDFLASLRTAKLSAEPVTDLSASDTQVAQENTLTVKNKEGEIHLMKDKAKHLDKC LKMLDMSFKDAERGDDTSCENLLDAFSIKLSETHGYGVQEEFTEENKLLEACIFKNNELL KNIQDVQSQISKIGLKDPTVPAVKHRKKSLIRLDKVLDEYEEEKRHLQEMANSLPHFKDG REKTVNQQCQNTVVLWENTKALVTECLEQCGRVLELLKQYQNFKSILTTLIQKEESVISL QASYMGKENLKKRIAEIEIVKEEFNEHLEVVDKINQVCKNLQFYLNKMKTFEEPPFEKEA NIIVDRWLDINEKTEDYYENLGRALALWDKLFNLKNVIDEWTEKALQKMELHQLTEEDRE RLKEELQVHEQKTSEFSRRVAEIQFLLQSSEIPLELQVMESSILNKMEHVQKCLTGESNC HALSGSTAELREDLDQAKTQIGMTESLLKALSPSDSLEIFTKLEEIQQQILQQKHSMILL ENQIGCLTPELSELKKQYESVSDLFNTKKSVLQDHFSKLLNDQCKNFNDWFSNIKVNLKE CFESSETKKSVEQKLQKLSDFLTLEGRNSKIKQVDSVLKHVKKHLPKAHVKELISWLVGQ EFELEKMESICQARAKELEDSLQQLLRLQDDHRNLRKWLTNQEEKWKGMEEPGEKTELFC QALARKREQFESVAQLNNSLKEYGFTEEEEIIMEATCLMDRYQTLLRQLSEIEEEDKLLP TEDQSFNDLAHDVIHWIKEIKESLMVLNSSEGKMPLEERIQKIKEIILLKPEGDARIETI MKQAESSEAPLVQKTLTDISNQWDNTLHLASTYLSHQEKLLLEGEKYLQSKEDLRLMLIE LKKKQEAGFALQHGLQEKKAQLKIYKKFLKKAQDLTSLLKELKSQGNYLLECTKNPSFSE EPWLEIKHLHESLLQQLQDSVQNLDGHVREHDSYQVCVTDLNTTLDNFSKEFVSFSDKPV DQIAVEEKLQKLQELENRLSLQDGTLKKILALAKSVKQNTSSVGQKIIKDDIKSLQCKQK DLENRLASAKQEMECCLNSILKSKRSTEKKGKFTLPGREKQATSDVQESTQESAAVEKLE EDWEINKDSAVEMAMSKQLSLNAQESMKNTEDERKVNELQNQPLELDTMLRNEQLEEIEK LYTQLEAKKAAIKPLEQTECLNKTETGALVLHNIGYSAQHLDNLLQALITLKKNKESQYC VLRDFQEYLAAVESSMKALLTDKESLKVGPLDSVTYLDKIKKFIASIEKEKDSLGNLKIK WENLSNHVTDMDKKLLESQIKQLEHGWEQVEQQIQKKYSQQVVEYDEFTTLMNKVQDTEI SLQQQQQHLQLRLKSPEERAGNQSMIALTTDLQATKHGFSVLKGQAELQMKRIWGEKEKK NLEDGINNLKKQWETLEPLHLEAENQIKKCDIRNKMKETILWAKNLLGELNPSIPLLPDD ILSQIRKCKVTHDGILARQQSVESLAEEVKDKVPSLTTYEGSDLNNTLEDLRNQYQMLVL KSTQRSQQLEFKLEERSNFFAIIRKFQLMVQESETLIIPRVETAATEAELKHHHVTLEAS QKELQEIDSGISTHLQELTNIYEELNVFERLFLEDQLKNLKIRTNRIQRFIQNTCNEVEH KIKFCRQFHEKTSALQEEADSIQRNELLLNQEVNKGVKEEIYNLKDRLTAIKCCILQVLK LKKVFDYIGLNWDFSQLDQLQTQVFEKEKELEEKIKQLDTFEEEHGKYQALLSKMRAIDL QIKKMTEVVLKAPDSSPESRRLNAQILSQRIEKAKCLCDEIIKKLNENKTFDDSFKEKEI LQIKLNAEENDKLYKVLQNMVLELSPKELDEKNCQDKLETSLHVLNQIKSQLQQPLLINL EIKHIQNEKDNCEAFQEQVWAEMCSIKAVTAIEKQREENSSEASDVETKLREFEDLQMQL NTSIDLRTNVLNDAYENLTRYKEAVTRAVESITSLEAIIIPYRVDVGNPEESLEMPLRKQ EELESTVAHIQDLTEKLGMISSPEAKLQLQYTLQELVSKNSAMKEAFKAQETEAERYLEN YKCYRKMEEDIYTNLSKMETVLGQSMSSLPLSYREALERLEQSKALVSNLISTKEELMKL RQILRLLRLRCTENDGICLLKIVSALWEKWLSLLEAAKEWEMWCEELKQEWKFVSEEIER EAIILDNLQEELPEISKTKEAATTEELSELLDCLCQYGENVEKQQLLLTLLLQRIRSIQN VPESSGAVETVPAFQEITSMKERCNKLLQKVQKNKELVQTEIQERHSFTKEIIALKNFFQ QTTTSFQNMAFQDHPEKSEQFEELQSILKKGKLTFENIMEKLRIKYSEMYTIVPAEIESQ VEECRKALEDIDEKISNEVLKSSPSYAMRRKIEEINNGLHNVEKMLQQKSKNIEKAQEIQ KKMWDELDLWHSKLNELDSEVQDIVEQDPGQAQEWMDNLMIPFQQYQQVSQRAECRTSQL NKATVKMEEYSDLLKSTEAWIENTSHLLANPADYDSLRTLSHHASTVQMALEDSEQKHNL LHSIFMDLEDLSIIFETDELTQSIQELSNQVTALQQKIMESLPQIQRMADDVVAIESEVK SMEKRVSKIKTILLSKEIFDFSPEEHLKHGEVILENIRPMKKTIAEIVSYQVELRLPQTG MKPLPVFQRTNQLLQDIKLLENVTQEQNELLKVVIKQTNEWDEEIENLKQILNNYSAQFS LEHMSPDQADKLPQLQGEIERMEKQILSLNQRKEDLLVDLKATVLNLHQHLKQEQEGVER DRLPAVTSEEGGVAERDASERKLNRRGSMSYLAAVEEEVEESSVKSDNGDEKAEPSPQSW SSLWKHDKDMEEDRASSSSGTIVQEAYGKISTSDNSMAQILTPDSLNTEQGPECSLRPNQ TEEGTTPPIEADTLDSSDAQGGLEPRVEKTRPEPTEVLHACKTQVAELELWLQQANVAVE PETLNADMQQVLEQQLVGCQAMLTEIEHKVAFLLETCKDQGLGDNGATQHEAEALSLKLK TVKCNLEKVQMMLQEKHSEDQHPTILKKSSEPEHQEALQPVNLSELESIVTERPQFSRQK DFQQQQVLELKPMEQKDFIKFIEFNAKKMWPQYCQHDNDTTQESSASNQASSPENDVPDS ILSPQGQNGDKWQYLHHELSSKIKLPLPQLVEPQVSTNMGILPSVTMYNFRYPTTEELKT YTTQLEDLRQEASNLQTQENMTEEAYINLDKKLFELFLTLSQCLSSVEEMLEMPRLYRED GSGQQVHYETLALELKKLYLALSDKKGDLLKAMTWPGENTNLLLECFDNLQVCLEHTQAA AVCRSKSLKAGLDYNRSYQNEIKRLYHQLIKSKTSLQQSLNEISGQSVAEQLQKADAYTV ELENAESRVAKLRDEGERLHLPYALLQEVYKLEDVLDSMWGMLRARYTELSSPFVTESQQ DALLQGMVELVKIGKEKLAHGHLKQTKSKVALQAQIENHKVFFQKLVADMLLIQAYSAKI LPSLLQNRETFWAEQVTEVKILEEKSRQCGMKLQSLLQKWEEFDENYASLEKDLEILIST LPSVSLVEETEERLVERISFYQQIKRNIGGKHARLYQTLNEGKQLVASVSCPELEGQIAK LEEQWLSLNKKIDHELHRLQALLKHLLSYNRDSDQLTKWLESSQHTLNYWKEQSLNVSQD LDTIRSNINNFFEFSKEVDEKSSLKTAVISIGNQLLHLKETDTATLRASLAQFEQKWTML ITQLPDIQEKLHQLQMEKLPSRKAITEMISWMNNVEHQTSDEDSVHSPSSASQVKHLLQK HKEFRMEMDYKQWIVDFVNQSLLQLSTCDVESKRYERTEFAEHLGEMNRQWHRVHGMLNR KIQHLEQLLESITESENKIQILNNWLEAQEERLKTLQKPESVISVQKLLLDCQDIENQLA IKSKALDELKQSYLTLESGAVPLLEDTASRIDELFQKRSSVLTQVNQLKTSMQSVLQEWK IYDQLYDEVNMMTIRFWYCMEHSKPVVLSLETLRCQVENLQSLQDEAESSEGSWEKLQEV IGKLKGLCPSVAEIIEEKCQNTHKRWTQVNQAIADQLQKAQSLLQLWKAYSNAHGEAAAR LKQQEAKFQQLANISMSGNNLAEILPPALQDIKELQHDVQKTKEAFLQNSSVLDRLPQPA ESSTHMLLPGPLHSLQRAAYLEKMLLVKANEFEFVLSQFKDFGVRLESLKGLIMHEEENL DRLHQQEKENPDSFLNHVLALTAQSPDIEHLNEVSLKLPLSDVAVKTLQNMNRQWIRATA TALERCSELQGIGLNEKFLYCCEKWIQLLEKIEEALKVDVANSLPELLEQQKTYKMLEAE VSINQTIADSYVTQSLQLLDTTEIENRPEFITEFSKLTDRWQNAVQGVRQRKGDVDGLVR QWQDFTTSVENLFRFLTDTSHLLSAVKGQERFSLYQTRSLIHELKNKEIHFQRRRTTCAL TLEAGEKLLLTTDLKTKESVGRRISQLQDSWKDMEPQLAEMIKQFQSTVETWDQCEKKIK ELKSRLQVLKAQSEDPLPELHEDLHNEKELIKELEQSLASWTQNLKELQTMKADLTRHVL VEDVMVLKEQIEHLHRQWEDLCLRVAIRKQEIEDRLNTWVVFNEKNKELCAWLVQMENKV LQTADISIEEMIEKLQKDCMEEINLFSENKLQLKQMGDQLIKASNKSRAAEIDDKLNKIN DRWQHLFDVIGSRVKKLKETFAFIQQLDKNMSNLRTWLARIESELSKPVVYDVCDDQEIQ KRLAEQQDLQRDIEQHSAGVESVFNICDVLLHDSDACANETECDSIQQTTRSLDRRWRNI CAMSMERRMKIEETWRLWQKFLDDYSRFEDWLKSAERTAACPNSSEVLYTSAKEELKRFE AFQRQIHERLTQLELINKQYRRLARENRTDTASRLKQMVHEGNQRWDNLQRRVTAVLRRL RHFTNQREEFEGTRESILVWLTEMDLQLTNVEHFSESDADDKMRQLNGFQQEITLNTNKI DQLIVFGEQLIQKSEPLDAVLIEDELEELHRYCQEVFGRVSRFHRRLTSCTPGLEDEKEA SENETDMEDPREIQTDSWRKRGESEEPSSPQSLCHLVAPGHERSGCETPVSVDSIPLEWD HTGDVGGSSSHEEDEEGPYYSALSGKSISDGHSWHVPDSPSCPEHHYKQMEGDRNVPPVP PASSTPYKPPYGKLLLPPGTDGGKEGPRVLNGNPQQEDGGLAGITEQQSGAFDRWEMIQA QELHNKLKIKQNLQQLNSDISAITTWLKKTEAELEMLKMAKPPSDIQEIELRVKRLQEIL KAFDTYKALVVSVNVSSKEFLQTESPESTELQSRLRQLSLLWEAAQGAVDSWRGGLRQSL MQCQDFHQLSQNLLLWLASAKNRRQKAHVTDPKADPRALLECRRELMQLEKELVERQPQV DMLQEISNSLLIKGHGEDCIEAEEKVHVIEKKLKQLREQVSQDLMALQGTQNPASPLPSF DEVDSGDQPPATSVPAPRAKQFRAVRTTEGEEETESRVPGSTRPQRSFLSRVVRAALPLQ LLLLLLLLLACLLPSSEEDYSCTQANNFARSFYPMLRYTNGPPPT |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Nesprin family
|
|||||
| Subcellular location |
Nucleus outer membrane; Cell junction, focal adhesion
|
|||||
| Function |
Multi-isomeric modular protein which forms a linking network between organelles and the actin cytoskeleton to maintain the subcellular spatial organization. As a component of the LINC (LInker of Nucleoskeleton and Cytoskeleton) complex involved in the connection between the nuclear lamina and the cytoskeleton. The nucleocytoplasmic interactions established by the LINC complex play an important role in the transmission of mechanical forces across the nuclear envelope and in nuclear movement and positioning. Specifically, SYNE2 and SUN2 assemble in arrays of transmembrane actin-associated nuclear (TAN) lines which are bound to F-actin cables and couple the nucleus to retrograde actin flow during actin-dependent nuclear movement. May be involved in nucleus-centrosome attachment. During interkinetic nuclear migration (INM) at G2 phase and nuclear migration in neural progenitors its LINC complex association with SUN1/2 and probable association with cytoplasmic dynein-dynactin motor complexes functions to pull the nucleus toward the centrosome; SYNE1 and SYNE2 may act redundantly. During INM at G1 phase mediates respective LINC complex association with kinesin to push the nucleus away from the centrosome. Involved in nuclear migration in retinal photoreceptor progenitors. Required for centrosome migration to the apical cell surface during early ciliogenesis. Facilitates the relaxation of mechanical stress imposed by compressive actin fibers at the rupture site through its nteraction with SYN2.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
DA-P2 Probe Info |
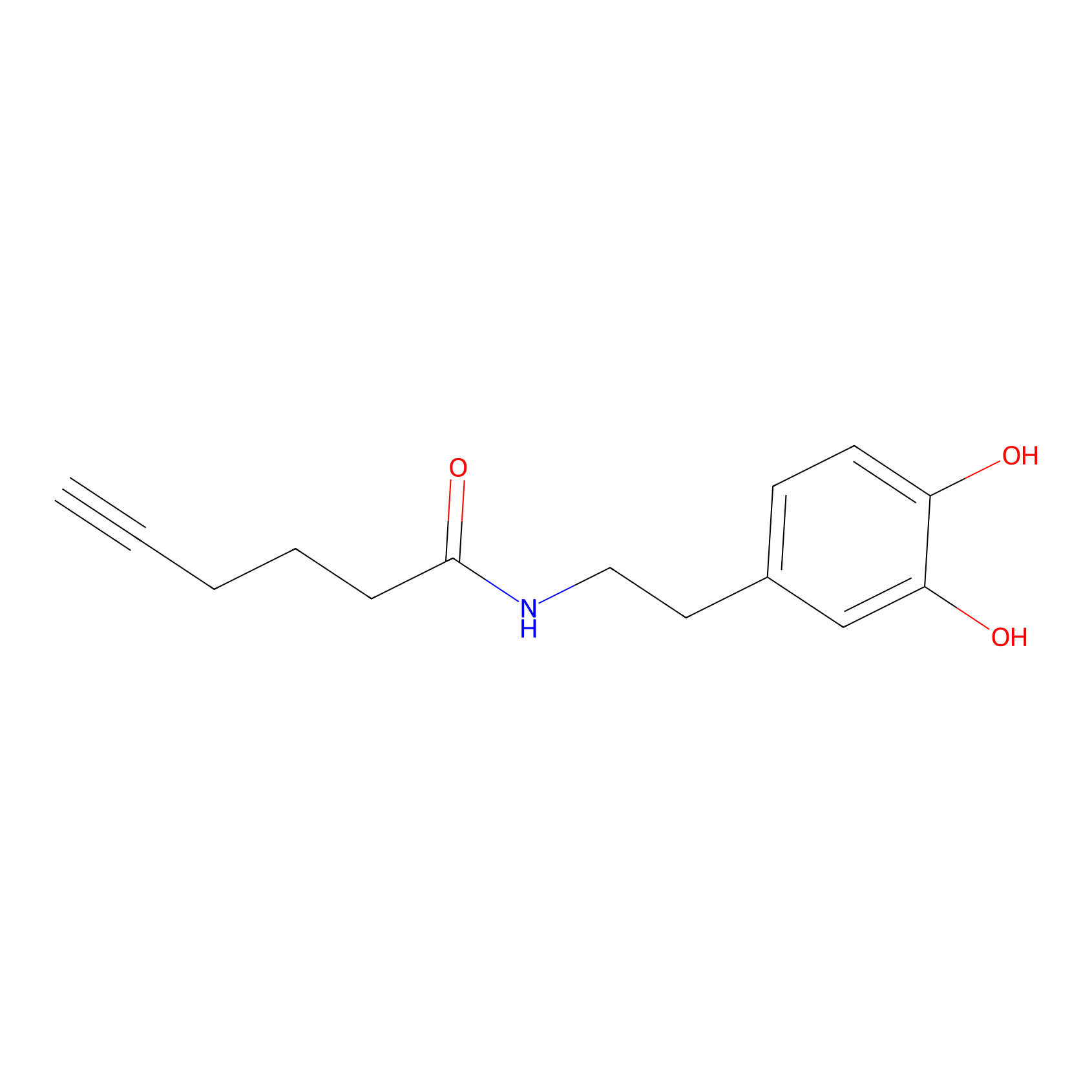 |
23.42 | LDD0348 | [2] | |
|
HRP Probe Info |
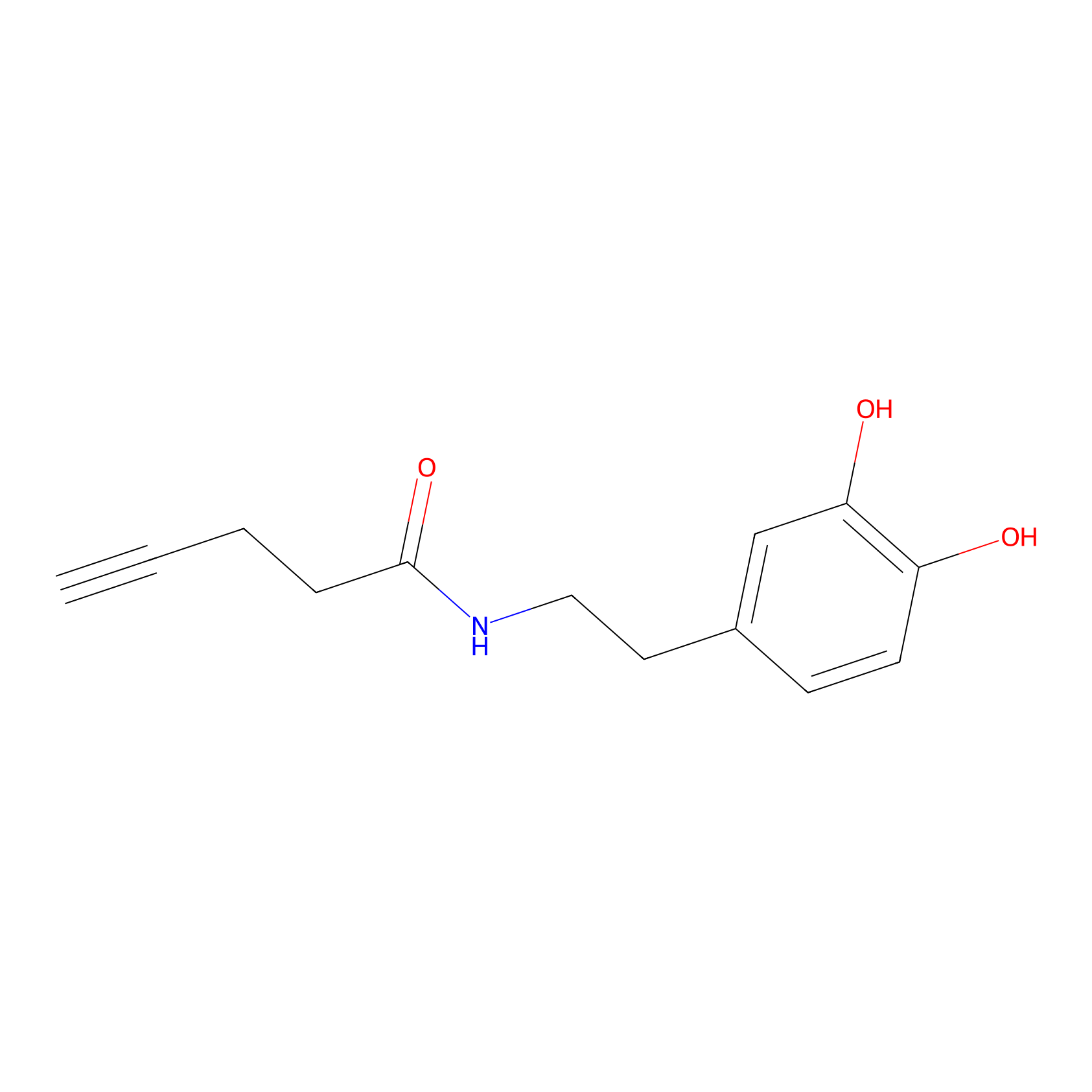 |
21.98 | LDD0347 | [2] | |
|
TH211 Probe Info |
 |
Y5390(11.45) | LDD0260 | [3] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [4] | |
|
OPA-S-S-alkyne Probe Info |
 |
K1618(1.41) | LDD3494 | [5] | |
|
Probe 1 Probe Info |
 |
Y1480(28.22); Y1638(13.28); Y3361(9.99); Y4678(5.01) | LDD3495 | [6] | |
|
BTD Probe Info |
 |
C2480(0.51) | LDD2089 | [7] | |
|
DA-P3 Probe Info |
 |
27.21 | LDD0185 | [2] | |
|
AHL-Pu-1 Probe Info |
 |
C553(2.06); C3513(2.50); C2480(2.24) | LDD0168 | [8] | |
|
DBIA Probe Info |
 |
C90(198.84); C1844(21.06); C2520(39.68) | LDD0209 | [9] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C39(0.00); C2935(0.00); C1432(0.00); C5212(0.00) | LDD0038 | [10] | |
|
IA-alkyne Probe Info |
 |
C601(0.00); C39(0.00); C1235(0.00); C1032(0.00) | LDD0036 | [10] | |
|
Lodoacetamide azide Probe Info |
 |
C2480(0.00); C601(0.00); C1235(0.00); C5586(0.00) | LDD0037 | [10] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [11] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [12] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [12] | |
|
ENE Probe Info |
 |
N.A. | LDD0006 | [13] | |
|
IPM Probe Info |
 |
C553(0.00); C2994(0.00); C90(0.00); C2480(0.00) | LDD0005 | [13] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [13] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [13] | |
|
VSF Probe Info |
 |
C2993(0.00); C5359(0.00); C553(0.00); C2480(0.00) | LDD0007 | [13] | |
|
Phosphinate-6 Probe Info |
 |
C1380(0.00); C6161(0.00) | LDD0018 | [14] | |
|
Acrolein Probe Info |
 |
C6739(0.00); C2257(0.00) | LDD0217 | [15] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [15] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [15] | |
|
AOyne Probe Info |
 |
8.40 | LDD0443 | [16] | |
|
NAIA_5 Probe Info |
 |
C1235(0.00); C5359(0.00); C5930(0.00); C3564(0.00) | LDD2223 | [17] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C2480(0.89) | LDD2112 | [7] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C1844(1.05) | LDD2117 | [7] |
| LDCM0025 | 4SU-RNA | HEK-293T | C553(2.06); C3513(2.50); C2480(2.24) | LDD0168 | [8] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C5930(3.21); C553(2.05) | LDD0169 | [8] |
| LDCM0214 | AC1 | HEK-293T | C90(0.97); C6739(0.93); C568(0.97); C1931(0.94) | LDD1507 | [18] |
| LDCM0215 | AC10 | HEK-293T | C90(1.01); C6739(0.97); C568(1.02); C1931(1.07) | LDD1508 | [18] |
| LDCM0226 | AC11 | HEK-293T | C90(1.02); C6739(0.99); C568(0.99); C1931(1.09) | LDD1509 | [18] |
| LDCM0237 | AC12 | HEK-293T | C90(0.99); C6739(0.97); C568(0.99); C1861(1.02) | LDD1510 | [18] |
| LDCM0259 | AC14 | HEK-293T | C90(1.05); C6739(1.20); C568(0.94); C1931(1.04) | LDD1512 | [18] |
| LDCM0270 | AC15 | HEK-293T | C90(0.96); C6739(1.16); C568(1.00); C1931(1.01) | LDD1513 | [18] |
| LDCM0276 | AC17 | HEK-293T | C90(0.99); C6739(0.94); C568(1.03); C1931(0.92) | LDD1515 | [18] |
| LDCM0277 | AC18 | HEK-293T | C90(1.02); C6739(0.91); C568(1.09); C1931(0.97) | LDD1516 | [18] |
| LDCM0278 | AC19 | HEK-293T | C90(0.97); C6739(0.91); C568(1.01); C1931(0.91) | LDD1517 | [18] |
| LDCM0279 | AC2 | HEK-293T | C90(0.99); C6739(0.94); C568(1.01); C1931(0.94) | LDD1518 | [18] |
| LDCM0280 | AC20 | HEK-293T | C90(1.01); C6739(1.24); C568(1.04); C1861(1.04) | LDD1519 | [18] |
| LDCM0281 | AC21 | HEK-293T | C90(0.94); C6739(0.98); C568(1.02); C1931(0.95) | LDD1520 | [18] |
| LDCM0282 | AC22 | HEK-293T | C90(0.92); C6739(1.00); C568(0.98); C1931(1.01) | LDD1521 | [18] |
| LDCM0283 | AC23 | HEK-293T | C90(0.98); C6739(0.99); C568(0.97); C1931(0.97) | LDD1522 | [18] |
| LDCM0284 | AC24 | HEK-293T | C90(0.94); C6739(1.03); C568(1.02); C1931(1.00) | LDD1523 | [18] |
| LDCM0285 | AC25 | HEK-293T | C90(0.98); C6739(0.95); C568(1.06); C1931(0.96) | LDD1524 | [18] |
| LDCM0286 | AC26 | HEK-293T | C90(1.06); C6739(0.98); C568(0.99); C1931(1.06) | LDD1525 | [18] |
| LDCM0287 | AC27 | HEK-293T | C90(1.01); C6739(1.07); C568(0.98); C1931(0.90) | LDD1526 | [18] |
| LDCM0288 | AC28 | HEK-293T | C90(0.99); C6739(0.93); C568(1.03); C1861(1.09) | LDD1527 | [18] |
| LDCM0289 | AC29 | HEK-293T | C90(0.97); C6739(1.00); C568(1.04); C1931(0.98) | LDD1528 | [18] |
| LDCM0290 | AC3 | HEK-293T | C90(1.03); C6739(1.02); C568(0.94); C1931(0.99) | LDD1529 | [18] |
| LDCM0291 | AC30 | HEK-293T | C90(0.99); C6739(0.88); C568(0.90); C1931(1.00) | LDD1530 | [18] |
| LDCM0292 | AC31 | HEK-293T | C90(0.93); C6739(1.01); C568(0.97); C1931(1.01) | LDD1531 | [18] |
| LDCM0293 | AC32 | HEK-293T | C90(0.99); C6739(1.02); C568(1.05); C1931(1.00) | LDD1532 | [18] |
| LDCM0294 | AC33 | HEK-293T | C90(1.02); C6739(0.96); C568(1.10); C1931(1.04) | LDD1533 | [18] |
| LDCM0295 | AC34 | HEK-293T | C90(0.96); C6739(1.02); C568(0.99); C1931(0.94) | LDD1534 | [18] |
| LDCM0296 | AC35 | HEK-293T | C90(0.95); C6739(1.06); C568(0.98); C1931(1.00) | LDD1535 | [18] |
| LDCM0297 | AC36 | HEK-293T | C90(1.07); C6739(1.01); C568(0.93); C1861(0.82) | LDD1536 | [18] |
| LDCM0298 | AC37 | HEK-293T | C90(1.02); C6739(0.95); C568(1.01); C1931(0.90) | LDD1537 | [18] |
| LDCM0299 | AC38 | HEK-293T | C90(0.96); C6739(1.00); C568(0.94); C1931(1.11) | LDD1538 | [18] |
| LDCM0300 | AC39 | HEK-293T | C90(0.99); C6739(1.15); C568(0.98); C1931(1.05) | LDD1539 | [18] |
| LDCM0301 | AC4 | HEK-293T | C90(1.05); C6739(0.96); C568(0.95); C1861(0.87) | LDD1540 | [18] |
| LDCM0302 | AC40 | HEK-293T | C90(0.99); C6739(0.95); C568(1.10); C1931(1.05) | LDD1541 | [18] |
| LDCM0303 | AC41 | HEK-293T | C90(0.96); C6739(1.07); C568(1.06); C1931(1.00) | LDD1542 | [18] |
| LDCM0304 | AC42 | HEK-293T | C90(1.02); C6739(0.90); C568(1.04); C1931(0.91) | LDD1543 | [18] |
| LDCM0305 | AC43 | HEK-293T | C90(1.07); C6739(0.96); C568(1.02); C1931(1.12) | LDD1544 | [18] |
| LDCM0306 | AC44 | HEK-293T | C90(0.94); C6739(1.04); C568(0.96); C1861(0.88) | LDD1545 | [18] |
| LDCM0307 | AC45 | HEK-293T | C90(1.05); C6739(0.95); C568(0.96); C1931(0.94) | LDD1546 | [18] |
| LDCM0308 | AC46 | HEK-293T | C90(1.04); C6739(0.88); C568(1.03); C1931(0.96) | LDD1547 | [18] |
| LDCM0309 | AC47 | HEK-293T | C90(0.97); C6739(1.19); C568(1.00); C1931(0.85) | LDD1548 | [18] |
| LDCM0310 | AC48 | HEK-293T | C90(1.03); C6739(1.03); C568(1.10); C1931(0.97) | LDD1549 | [18] |
| LDCM0311 | AC49 | HEK-293T | C90(0.94); C6739(1.11); C568(1.04); C1931(0.99) | LDD1550 | [18] |
| LDCM0312 | AC5 | HEK-293T | C90(1.03); C6739(0.97); C568(0.97); C1931(0.94) | LDD1551 | [18] |
| LDCM0313 | AC50 | HEK-293T | C90(0.98); C6739(1.01); C568(1.06); C1931(0.95) | LDD1552 | [18] |
| LDCM0314 | AC51 | HEK-293T | C90(1.02); C6739(0.94); C568(0.95); C1931(0.96) | LDD1553 | [18] |
| LDCM0315 | AC52 | HEK-293T | C90(0.98); C6739(1.01); C568(0.98); C1861(0.91) | LDD1554 | [18] |
| LDCM0316 | AC53 | HEK-293T | C90(1.00); C6739(1.03); C568(1.01); C1931(0.94) | LDD1555 | [18] |
| LDCM0317 | AC54 | HEK-293T | C90(0.96); C6739(1.09); C568(0.92); C1931(0.99) | LDD1556 | [18] |
| LDCM0318 | AC55 | HEK-293T | C90(0.96); C6739(1.19); C568(0.98); C1931(0.98) | LDD1557 | [18] |
| LDCM0319 | AC56 | HEK-293T | C90(0.98); C6739(0.94); C568(1.04); C1931(0.99) | LDD1558 | [18] |
| LDCM0320 | AC57 | HEK-293T | C90(1.00); C6739(1.00); C568(1.01); C1931(0.90) | LDD1559 | [18] |
| LDCM0321 | AC58 | HEK-293T | C90(0.99); C6739(0.99); C568(1.02); C1931(1.02) | LDD1560 | [18] |
| LDCM0322 | AC59 | HEK-293T | C90(1.01); C6739(0.90); C568(0.97); C1931(0.94) | LDD1561 | [18] |
| LDCM0323 | AC6 | HEK-293T | C90(1.00); C6739(1.03); C568(0.93); C1931(1.07) | LDD1562 | [18] |
| LDCM0324 | AC60 | HEK-293T | C90(1.01); C6739(1.08); C568(1.00); C1861(1.00) | LDD1563 | [18] |
| LDCM0325 | AC61 | HEK-293T | C90(1.05); C6739(1.01); C568(1.05); C1931(0.94) | LDD1564 | [18] |
| LDCM0326 | AC62 | HEK-293T | C90(0.98); C6739(1.11); C568(0.96); C1931(1.05) | LDD1565 | [18] |
| LDCM0327 | AC63 | HEK-293T | C90(0.97); C6739(1.00); C568(1.07); C1931(1.02) | LDD1566 | [18] |
| LDCM0328 | AC64 | HEK-293T | C90(1.00); C6739(0.96); C568(1.06); C1931(0.98) | LDD1567 | [18] |
| LDCM0334 | AC7 | HEK-293T | C90(1.02); C6739(1.12); C568(0.96); C1931(1.02) | LDD1568 | [18] |
| LDCM0345 | AC8 | HEK-293T | C90(1.02); C6739(0.96); C568(1.05); C1931(0.96) | LDD1569 | [18] |
| LDCM0248 | AKOS034007472 | HEK-293T | C90(1.07); C6739(0.99); C568(1.01); C1931(0.96) | LDD1511 | [18] |
| LDCM0356 | AKOS034007680 | HEK-293T | C90(1.03); C6739(0.97); C568(1.02); C1931(1.03) | LDD1570 | [18] |
| LDCM0275 | AKOS034007705 | HEK-293T | C90(1.05); C6739(1.00); C568(1.04); C1931(0.96) | LDD1514 | [18] |
| LDCM0087 | Capsaicin | HEK-293T | 27.21 | LDD0185 | [2] |
| LDCM0108 | Chloroacetamide | HeLa | C5212(0.00); C2257(0.00); C6702(0.00) | LDD0222 | [15] |
| LDCM0632 | CL-Sc | Hep-G2 | C1151(20.00); C39(2.56); C1980(1.50); C2935(1.11) | LDD2227 | [17] |
| LDCM0367 | CL1 | HEK-293T | C90(1.02); C568(1.37); C1931(1.04); C1861(1.01) | LDD1571 | [18] |
| LDCM0368 | CL10 | HEK-293T | C90(1.04); C6739(0.85); C568(3.05); C1931(0.79) | LDD1572 | [18] |
| LDCM0369 | CL100 | HEK-293T | C90(0.90); C6739(1.00); C568(1.22); C1931(0.97) | LDD1573 | [18] |
| LDCM0370 | CL101 | HEK-293T | C90(1.02); C568(1.09); C1931(0.97); C1861(1.10) | LDD1574 | [18] |
| LDCM0371 | CL102 | HEK-293T | C90(1.02); C6739(0.91); C568(1.69); C1931(0.98) | LDD1575 | [18] |
| LDCM0372 | CL103 | HEK-293T | C90(0.95); C6739(1.06); C568(1.12); C1931(1.00) | LDD1576 | [18] |
| LDCM0373 | CL104 | HEK-293T | C90(0.92); C6739(1.02); C568(1.23); C1931(1.27) | LDD1577 | [18] |
| LDCM0374 | CL105 | HEK-293T | C90(0.98); C568(1.16); C1931(0.98); C1861(0.96) | LDD1578 | [18] |
| LDCM0375 | CL106 | HEK-293T | C90(1.01); C6739(0.93); C568(1.22); C1931(1.00) | LDD1579 | [18] |
| LDCM0376 | CL107 | HEK-293T | C90(0.98); C6739(0.98); C568(0.95); C1931(1.00) | LDD1580 | [18] |
| LDCM0377 | CL108 | HEK-293T | C90(0.89); C6739(1.06); C568(1.15); C1931(1.00) | LDD1581 | [18] |
| LDCM0378 | CL109 | HEK-293T | C90(1.05); C568(1.24); C1931(0.93); C1861(1.05) | LDD1582 | [18] |
| LDCM0379 | CL11 | HEK-293T | C90(0.97); C6739(1.02); C568(1.58); C1931(1.04) | LDD1583 | [18] |
| LDCM0380 | CL110 | HEK-293T | C90(0.97); C6739(0.87); C568(2.03); C1931(1.00) | LDD1584 | [18] |
| LDCM0381 | CL111 | HEK-293T | C90(1.01); C6739(0.96); C568(1.54); C1931(1.00) | LDD1585 | [18] |
| LDCM0382 | CL112 | HEK-293T | C90(0.92); C6739(0.98); C568(1.18); C1931(1.08) | LDD1586 | [18] |
| LDCM0383 | CL113 | HEK-293T | C90(0.93); C568(0.96); C1931(1.02); C1861(0.96) | LDD1587 | [18] |
| LDCM0384 | CL114 | HEK-293T | C90(0.98); C6739(0.91); C568(2.13); C1931(0.99) | LDD1588 | [18] |
| LDCM0385 | CL115 | HEK-293T | C90(0.95); C6739(1.01); C568(1.03); C1931(1.04) | LDD1589 | [18] |
| LDCM0386 | CL116 | HEK-293T | C90(0.86); C6739(1.09); C568(1.00); C1931(1.03) | LDD1590 | [18] |
| LDCM0387 | CL117 | HEK-293T | C90(1.02); C568(0.98); C1931(0.97); C1861(0.97) | LDD1591 | [18] |
| LDCM0388 | CL118 | HEK-293T | C90(1.02); C6739(1.02); C568(0.95); C1931(1.05) | LDD1592 | [18] |
| LDCM0389 | CL119 | HEK-293T | C90(0.94); C6739(1.03); C568(0.99); C1931(1.12) | LDD1593 | [18] |
| LDCM0390 | CL12 | HEK-293T | C90(1.08); C6739(0.97); C568(1.27); C1931(1.03) | LDD1594 | [18] |
| LDCM0391 | CL120 | HEK-293T | C90(0.95); C6739(1.01); C568(1.08); C1931(1.08) | LDD1595 | [18] |
| LDCM0392 | CL121 | HEK-293T | C90(1.00); C568(1.04); C1931(0.90); C1861(1.16) | LDD1596 | [18] |
| LDCM0393 | CL122 | HEK-293T | C90(0.94); C6739(1.00); C568(1.25); C1931(0.99) | LDD1597 | [18] |
| LDCM0394 | CL123 | HEK-293T | C90(0.93); C6739(0.93); C568(2.22); C1931(0.95) | LDD1598 | [18] |
| LDCM0395 | CL124 | HEK-293T | C90(0.91); C6739(1.05); C568(1.38); C1931(1.20) | LDD1599 | [18] |
| LDCM0396 | CL125 | HEK-293T | C90(1.02); C568(1.04); C1931(0.95); C1861(1.11) | LDD1600 | [18] |
| LDCM0397 | CL126 | HEK-293T | C90(1.06); C6739(0.95); C568(1.21); C1931(0.95) | LDD1601 | [18] |
| LDCM0398 | CL127 | HEK-293T | C90(0.98); C6739(1.05); C568(1.01); C1931(0.96) | LDD1602 | [18] |
| LDCM0399 | CL128 | HEK-293T | C90(0.96); C6739(1.02); C568(1.00); C1931(0.85) | LDD1603 | [18] |
| LDCM0400 | CL13 | HEK-293T | C90(0.98); C568(1.12); C1931(0.98); C1861(1.04) | LDD1604 | [18] |
| LDCM0401 | CL14 | HEK-293T | C90(1.01); C6739(0.96); C568(0.96); C1931(1.06) | LDD1605 | [18] |
| LDCM0402 | CL15 | HEK-293T | C90(0.97); C6739(0.97); C568(2.35); C1931(1.09) | LDD1606 | [18] |
| LDCM0403 | CL16 | HEK-293T | C90(0.88); C6739(1.11); C568(1.02); C1931(1.04) | LDD1607 | [18] |
| LDCM0404 | CL17 | HEK-293T | C90(0.95); C6739(0.97); C568(2.15); C1931(0.95) | LDD1608 | [18] |
| LDCM0405 | CL18 | HEK-293T | C90(1.04); C6739(1.12); C568(1.05); C1931(1.04) | LDD1609 | [18] |
| LDCM0406 | CL19 | HEK-293T | C90(1.05); C6739(1.07); C568(1.01); C1931(1.07) | LDD1610 | [18] |
| LDCM0407 | CL2 | HEK-293T | C90(1.01); C6739(1.08); C568(0.97); C1931(1.04) | LDD1611 | [18] |
| LDCM0408 | CL20 | HEK-293T | C90(1.21); C6739(1.04); C568(1.06); C1861(1.06) | LDD1612 | [18] |
| LDCM0409 | CL21 | HEK-293T | C90(1.12); C6739(0.99); C568(2.32); C1931(0.94) | LDD1613 | [18] |
| LDCM0410 | CL22 | HEK-293T | C90(1.07); C6739(1.29); C568(1.12); C1931(1.11) | LDD1614 | [18] |
| LDCM0411 | CL23 | HEK-293T | C90(1.03); C6739(1.06); C568(1.12); C1931(1.04) | LDD1615 | [18] |
| LDCM0412 | CL24 | HEK-293T | C90(1.03); C6739(1.04); C568(1.11); C1931(1.15) | LDD1616 | [18] |
| LDCM0413 | CL25 | HEK-293T | C90(1.02); C568(1.47); C1931(0.94); C1861(1.17) | LDD1617 | [18] |
| LDCM0414 | CL26 | HEK-293T | C90(0.97); C6739(1.01); C568(0.97); C1931(1.05) | LDD1618 | [18] |
| LDCM0415 | CL27 | HEK-293T | C90(0.97); C6739(0.97); C568(1.04); C1931(1.06) | LDD1619 | [18] |
| LDCM0416 | CL28 | HEK-293T | C90(0.96); C6739(1.01); C568(1.22); C1931(1.09) | LDD1620 | [18] |
| LDCM0417 | CL29 | HEK-293T | C90(1.02); C6739(1.02); C568(1.08); C1931(1.12) | LDD1621 | [18] |
| LDCM0418 | CL3 | HEK-293T | C90(0.94); C6739(1.05); C568(1.15); C1931(0.98) | LDD1622 | [18] |
| LDCM0419 | CL30 | HEK-293T | C90(1.05); C6739(0.97); C568(0.99); C1931(1.03) | LDD1623 | [18] |
| LDCM0420 | CL31 | HEK-293T | C90(1.03); C6739(1.14); C568(1.14); C1931(0.87) | LDD1624 | [18] |
| LDCM0421 | CL32 | HEK-293T | C90(1.08); C6739(1.10); C568(1.16); C1861(1.16) | LDD1625 | [18] |
| LDCM0422 | CL33 | HEK-293T | C90(1.13); C6739(0.94); C568(3.90); C1931(0.89) | LDD1626 | [18] |
| LDCM0423 | CL34 | HEK-293T | C90(1.05); C6739(1.06); C568(1.58); C1931(1.04) | LDD1627 | [18] |
| LDCM0424 | CL35 | HEK-293T | C90(1.09); C6739(1.05); C568(1.11); C1931(1.31) | LDD1628 | [18] |
| LDCM0425 | CL36 | HEK-293T | C90(1.08); C6739(1.04); C568(1.45); C1931(1.05) | LDD1629 | [18] |
| LDCM0426 | CL37 | HEK-293T | C90(1.00); C568(1.10); C1931(1.02); C1861(1.09) | LDD1630 | [18] |
| LDCM0428 | CL39 | HEK-293T | C90(0.97); C6739(0.94); C568(1.46); C1931(0.97) | LDD1632 | [18] |
| LDCM0429 | CL4 | HEK-293T | C90(0.93); C6739(1.07); C568(1.58); C1931(1.02) | LDD1633 | [18] |
| LDCM0430 | CL40 | HEK-293T | C90(1.03); C6739(1.04); C568(1.01); C1931(1.14) | LDD1634 | [18] |
| LDCM0431 | CL41 | HEK-293T | C90(0.98); C6739(0.90); C568(1.42); C1931(0.94) | LDD1635 | [18] |
| LDCM0432 | CL42 | HEK-293T | C90(1.08); C6739(1.06); C568(1.02); C1931(1.04) | LDD1636 | [18] |
| LDCM0433 | CL43 | HEK-293T | C90(1.07); C6739(1.17); C568(1.12); C1931(0.95) | LDD1637 | [18] |
| LDCM0434 | CL44 | HEK-293T | C90(1.13); C6739(1.16); C568(1.13); C1861(1.15) | LDD1638 | [18] |
| LDCM0435 | CL45 | HEK-293T | C90(1.09); C6739(1.00); C568(1.96); C1931(0.90) | LDD1639 | [18] |
| LDCM0436 | CL46 | HEK-293T | C90(1.11); C6739(1.06); C568(1.19); C1931(1.01) | LDD1640 | [18] |
| LDCM0437 | CL47 | HEK-293T | C90(1.07); C6739(1.00); C568(1.20); C1931(1.05) | LDD1641 | [18] |
| LDCM0438 | CL48 | HEK-293T | C90(1.13); C6739(1.02); C568(1.22); C1931(1.13) | LDD1642 | [18] |
| LDCM0439 | CL49 | HEK-293T | C90(1.03); C568(1.02); C1931(1.01); C1861(1.10) | LDD1643 | [18] |
| LDCM0440 | CL5 | HEK-293T | C90(0.92); C6739(1.14); C568(0.97); C1931(0.96) | LDD1644 | [18] |
| LDCM0441 | CL50 | HEK-293T | C90(1.07); C6739(0.97); C568(1.47); C1931(1.02) | LDD1645 | [18] |
| LDCM0443 | CL52 | HEK-293T | C90(0.91); C6739(1.07); C568(1.08); C1931(1.05) | LDD1646 | [18] |
| LDCM0444 | CL53 | HEK-293T | C90(0.97); C6739(1.01); C568(2.08); C1931(1.07) | LDD1647 | [18] |
| LDCM0445 | CL54 | HEK-293T | C90(1.04); C6739(0.99); C568(2.78); C1931(0.96) | LDD1648 | [18] |
| LDCM0446 | CL55 | HEK-293T | C90(1.03); C6739(1.36); C568(1.07); C1931(0.94) | LDD1649 | [18] |
| LDCM0447 | CL56 | HEK-293T | C90(1.06); C6739(1.23); C568(1.08); C1861(0.99) | LDD1650 | [18] |
| LDCM0448 | CL57 | HEK-293T | C90(1.02); C6739(0.88); C568(2.58); C1931(0.93) | LDD1651 | [18] |
| LDCM0449 | CL58 | HEK-293T | C90(1.06); C6739(1.06); C568(1.85); C1931(1.00) | LDD1652 | [18] |
| LDCM0450 | CL59 | HEK-293T | C90(1.08); C6739(1.17); C568(1.27); C1931(0.99) | LDD1653 | [18] |
| LDCM0451 | CL6 | HEK-293T | C90(1.03); C6739(1.08); C568(2.40); C1931(1.07) | LDD1654 | [18] |
| LDCM0452 | CL60 | HEK-293T | C90(1.11); C6739(1.03); C568(1.33); C1931(1.15) | LDD1655 | [18] |
| LDCM0453 | CL61 | HEK-293T | C90(1.03); C568(0.95); C1931(0.91); C1861(0.98) | LDD1656 | [18] |
| LDCM0454 | CL62 | HEK-293T | C90(1.10); C6739(1.01); C568(0.97); C1931(0.97) | LDD1657 | [18] |
| LDCM0455 | CL63 | HEK-293T | C90(1.04); C6739(1.07); C568(0.99); C1931(1.19) | LDD1658 | [18] |
| LDCM0456 | CL64 | HEK-293T | C90(0.91); C6739(0.96); C568(2.03); C1931(0.88) | LDD1659 | [18] |
| LDCM0457 | CL65 | HEK-293T | C90(0.95); C6739(1.06); C568(0.98); C1931(0.97) | LDD1660 | [18] |
| LDCM0458 | CL66 | HEK-293T | C90(1.06); C6739(1.02); C568(1.21); C1931(1.05) | LDD1661 | [18] |
| LDCM0459 | CL67 | HEK-293T | C90(1.04); C6739(1.07); C568(1.01); C1931(1.20) | LDD1662 | [18] |
| LDCM0460 | CL68 | HEK-293T | C90(1.02); C6739(1.11); C568(1.22); C1861(1.01) | LDD1663 | [18] |
| LDCM0461 | CL69 | HEK-293T | C90(1.06); C6739(1.02); C568(1.68); C1931(1.06) | LDD1664 | [18] |
| LDCM0462 | CL7 | HEK-293T | C90(1.11); C6739(0.96); C568(1.10); C1931(0.99) | LDD1665 | [18] |
| LDCM0463 | CL70 | HEK-293T | C90(1.11); C6739(1.34); C568(1.05); C1931(1.04) | LDD1666 | [18] |
| LDCM0464 | CL71 | HEK-293T | C90(1.04); C6739(1.16); C568(1.16); C1931(0.91) | LDD1667 | [18] |
| LDCM0465 | CL72 | HEK-293T | C90(1.10); C6739(0.97); C568(1.08); C1931(1.11) | LDD1668 | [18] |
| LDCM0466 | CL73 | HEK-293T | C90(1.03); C568(1.20); C1931(0.98); C1861(1.10) | LDD1669 | [18] |
| LDCM0467 | CL74 | HEK-293T | C90(1.05); C6739(1.05); C568(0.88); C1931(1.01) | LDD1670 | [18] |
| LDCM0469 | CL76 | HEK-293T | C90(0.97); C6739(1.08); C568(1.05); C1931(1.03) | LDD1672 | [18] |
| LDCM0470 | CL77 | HEK-293T | C90(0.93); C6739(0.94); C568(1.71); C1931(0.88) | LDD1673 | [18] |
| LDCM0471 | CL78 | HEK-293T | C90(1.00); C6739(1.01); C568(1.04); C1931(1.05) | LDD1674 | [18] |
| LDCM0472 | CL79 | HEK-293T | C90(1.04); C6739(1.11); C568(1.05); C1931(1.00) | LDD1675 | [18] |
| LDCM0473 | CL8 | HEK-293T | C90(1.34); C6739(0.72); C568(2.96); C1861(1.39) | LDD1676 | [18] |
| LDCM0474 | CL80 | HEK-293T | C90(1.07); C6739(0.93); C568(1.02); C1861(1.25) | LDD1677 | [18] |
| LDCM0475 | CL81 | HEK-293T | C90(1.10); C6739(1.10); C568(1.06); C1931(1.05) | LDD1678 | [18] |
| LDCM0476 | CL82 | HEK-293T | C90(1.03); C6739(1.16); C568(1.05); C1931(1.04) | LDD1679 | [18] |
| LDCM0477 | CL83 | HEK-293T | C90(1.00); C6739(1.08); C568(1.48); C1931(1.13) | LDD1680 | [18] |
| LDCM0478 | CL84 | HEK-293T | C90(1.12); C6739(1.02); C568(1.63); C1931(1.06) | LDD1681 | [18] |
| LDCM0479 | CL85 | HEK-293T | C90(1.05); C568(0.98); C1931(0.96); C1861(1.02) | LDD1682 | [18] |
| LDCM0480 | CL86 | HEK-293T | C90(1.04); C6739(1.05); C568(1.07); C1931(1.07) | LDD1683 | [18] |
| LDCM0481 | CL87 | HEK-293T | C90(0.94); C6739(0.98); C568(1.11); C1931(0.90) | LDD1684 | [18] |
| LDCM0482 | CL88 | HEK-293T | C90(0.95); C6739(1.12); C568(1.03); C1931(1.17) | LDD1685 | [18] |
| LDCM0483 | CL89 | HEK-293T | C90(1.01); C6739(1.11); C568(1.08); C1931(0.88) | LDD1686 | [18] |
| LDCM0484 | CL9 | HEK-293T | C90(1.07); C6739(1.06); C568(1.34); C1931(0.96) | LDD1687 | [18] |
| LDCM0485 | CL90 | HEK-293T | C90(1.10); C6739(0.79); C568(3.45); C1931(0.78) | LDD1688 | [18] |
| LDCM0486 | CL91 | HEK-293T | C90(1.08); C6739(1.18); C568(1.03); C1931(1.16) | LDD1689 | [18] |
| LDCM0487 | CL92 | HEK-293T | C90(1.07); C6739(1.04); C568(1.59); C1861(0.80) | LDD1690 | [18] |
| LDCM0488 | CL93 | HEK-293T | C90(1.03); C6739(0.97); C568(2.28); C1931(0.91) | LDD1691 | [18] |
| LDCM0489 | CL94 | HEK-293T | C90(1.04); C6739(1.10); C568(1.15); C1931(1.16) | LDD1692 | [18] |
| LDCM0490 | CL95 | HEK-293T | C90(1.06); C6739(1.03); C568(5.16); C1931(1.08) | LDD1693 | [18] |
| LDCM0491 | CL96 | HEK-293T | C90(1.03); C6739(0.88); C568(1.58); C1931(1.01) | LDD1694 | [18] |
| LDCM0492 | CL97 | HEK-293T | C90(0.95); C568(1.45); C1931(0.92); C1861(1.10) | LDD1695 | [18] |
| LDCM0493 | CL98 | HEK-293T | C90(1.00); C6739(0.94); C568(1.28); C1931(1.04) | LDD1696 | [18] |
| LDCM0494 | CL99 | HEK-293T | C90(0.94); C6739(1.01); C568(1.20); C1931(0.99) | LDD1697 | [18] |
| LDCM0198 | Dimethyl Fumarate(DMF) | T cell | C1032(9.88); C568(15.97) | LDD0513 | [19] |
| LDCM0495 | E2913 | HEK-293T | C90(0.93); C6739(1.01); C568(1.22); C1931(0.89) | LDD1698 | [18] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C1151(3.24); C2480(3.21) | LDD1702 | [7] |
| LDCM0625 | F8 | Ramos | C1151(1.19); C90(1.23); C553(0.58); C39(0.87) | LDD2187 | [20] |
| LDCM0572 | Fragment10 | Ramos | C1151(1.39); C90(2.86); C1003(0.92); C39(2.26) | LDD2189 | [20] |
| LDCM0573 | Fragment11 | Ramos | C1151(15.29); C90(0.82); C553(6.34); C39(0.02) | LDD2190 | [20] |
| LDCM0574 | Fragment12 | Ramos | C1151(0.84); C90(4.90); C1003(1.06); C39(4.24) | LDD2191 | [20] |
| LDCM0575 | Fragment13 | Ramos | C1151(1.03); C90(0.95); C1003(0.70); C39(0.91) | LDD2192 | [20] |
| LDCM0576 | Fragment14 | Ramos | C1151(1.20); C39(2.45); C1235(1.77); C6161(0.83) | LDD2193 | [20] |
| LDCM0579 | Fragment20 | Ramos | C1151(0.83) | LDD2194 | [20] |
| LDCM0580 | Fragment21 | Ramos | C1151(1.01); C90(1.14); C1003(0.94); C553(0.97) | LDD2195 | [20] |
| LDCM0582 | Fragment23 | Ramos | C1151(1.25); C90(1.12); C553(0.90); C39(0.82) | LDD2196 | [20] |
| LDCM0578 | Fragment27 | Ramos | C1151(0.91); C90(0.85); C1003(1.00); C553(0.75) | LDD2197 | [20] |
| LDCM0586 | Fragment28 | Ramos | C1151(0.66); C90(0.39); C1003(0.78); C553(0.70) | LDD2198 | [20] |
| LDCM0588 | Fragment30 | Ramos | C1151(1.15); C90(1.17); C553(1.48); C39(0.82) | LDD2199 | [20] |
| LDCM0589 | Fragment31 | Ramos | C1151(0.93); C90(1.33); C1003(1.03); C39(1.10) | LDD2200 | [20] |
| LDCM0590 | Fragment32 | Ramos | C1151(1.44); C90(3.41); C1003(0.86); C39(3.78) | LDD2201 | [20] |
| LDCM0468 | Fragment33 | HEK-293T | C90(0.93); C6739(1.01); C568(1.12); C1931(0.96) | LDD1671 | [18] |
| LDCM0596 | Fragment38 | Ramos | C1151(0.87); C90(1.67); C1003(1.05); C39(1.08) | LDD2203 | [20] |
| LDCM0566 | Fragment4 | Ramos | C1151(1.01); C90(1.93); C1003(0.62); C39(1.81) | LDD2184 | [20] |
| LDCM0427 | Fragment51 | HEK-293T | C90(1.06); C6739(1.07); C568(0.99); C1931(1.09) | LDD1631 | [18] |
| LDCM0610 | Fragment52 | Ramos | C1151(0.85); C90(1.57); C1003(1.39); C39(1.05) | LDD2204 | [20] |
| LDCM0614 | Fragment56 | Ramos | C1151(1.44); C90(1.30); C553(0.69); C39(0.67) | LDD2205 | [20] |
| LDCM0569 | Fragment7 | Ramos | C1151(1.16); C90(2.66); C39(2.30); C1235(1.15) | LDD2186 | [20] |
| LDCM0571 | Fragment9 | Ramos | C1151(1.57); C90(1.31) | LDD2188 | [20] |
| LDCM0022 | KB02 | HEK-293T | C6739(0.91); C568(0.59); C1931(0.87); C1861(0.85) | LDD1492 | [18] |
| LDCM0023 | KB03 | Jurkat | C90(198.84); C1844(21.06); C2520(39.68) | LDD0209 | [9] |
| LDCM0024 | KB05 | COLO792 | C1598(1.82) | LDD3310 | [21] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0230 | [15] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C2480(0.51) | LDD2089 | [7] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C4324(1.16) | LDD2106 | [7] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C2480(0.97) | LDD2109 | [7] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C4324(0.68) | LDD2114 | [7] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C1598(1.64) | LDD2123 | [7] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C1598(1.22) | LDD2127 | [7] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C4324(0.75) | LDD2129 | [7] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C1598(1.42) | LDD2137 | [7] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C2480(0.79) | LDD2140 | [7] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C4324(0.69) | LDD2141 | [7] |
| LDCM0170 | Structure8 | Ramos | 20.00 | LDD0433 | [22] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| SUN domain-containing protein 1 (SUN1) | . | O94901 | |||
| SUN domain-containing protein 2 (SUN2) | . | Q9UH99 | |||
Other
References
