Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
Alkylaryl probe 3 Probe Info |
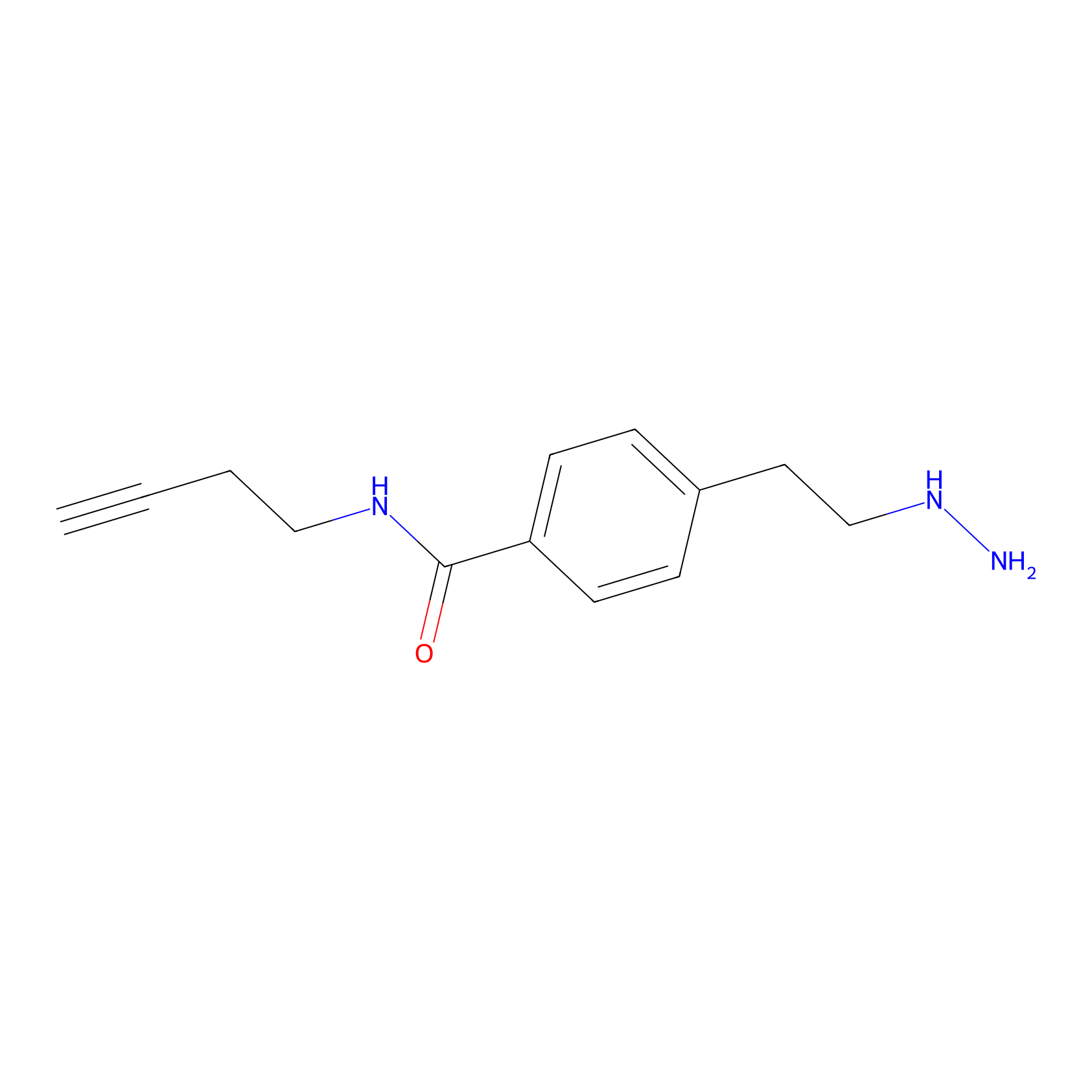 |
20.00 | LDD0381 | [1] | |
|
W1 Probe Info |
 |
10.64 | LDD0235 | [2] | |
|
YN-1 Probe Info |
 |
45.43 | LDD0444 | [3] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [3] | |
|
DBIA Probe Info |
 |
C370(1.71) | LDD3312 | [4] | |
|
AZ-9 Probe Info |
 |
10.00 | LDD2154 | [5] | |
|
IPM Probe Info |
 |
C186(14.14); C370(8.82); C133(4.42) | LDD1701 | [6] | |
|
IA-alkyne Probe Info |
 |
C370(0.00); C163(0.00); C186(0.00); C126(0.00) | LDD0162 | [7] | |
|
BTD Probe Info |
 |
C50(0.00); C370(0.00) | LDD0004 | [8] | |
|
WYneN Probe Info |
 |
C133(0.00); C126(0.00); C370(0.00) | LDD0021 | [8] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [8] | |
|
NHS Probe Info |
 |
K412(0.00); K362(0.00) | LDD0010 | [8] | |
|
HHS-475 Probe Info |
 |
Y115(1.05); Y119(1.57); Y132(1.27); Y154(0.89) | LDD2238 | [9] | |
|
HHS-482 Probe Info |
 |
Y115(0.87); Y119(1.08); Y132(0.80); Y297(1.08) | LDD2239 | [9] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
PARPYnD Probe Info |
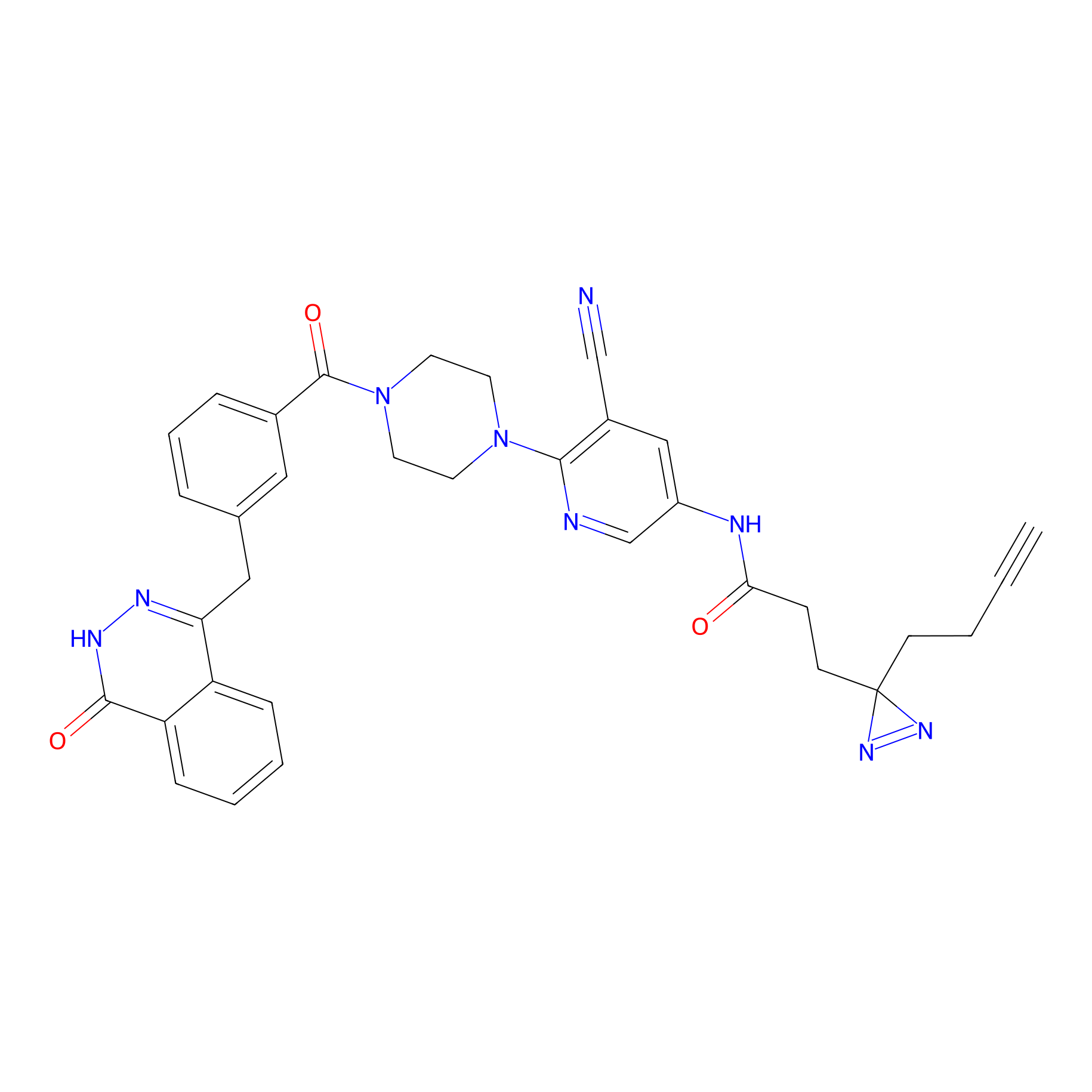 |
2.03 | LDD0374 | [10] | |
|
FFF probe13 Probe Info |
 |
5.59 | LDD0476 | [11] | |
|
AEA-DA Probe Info |
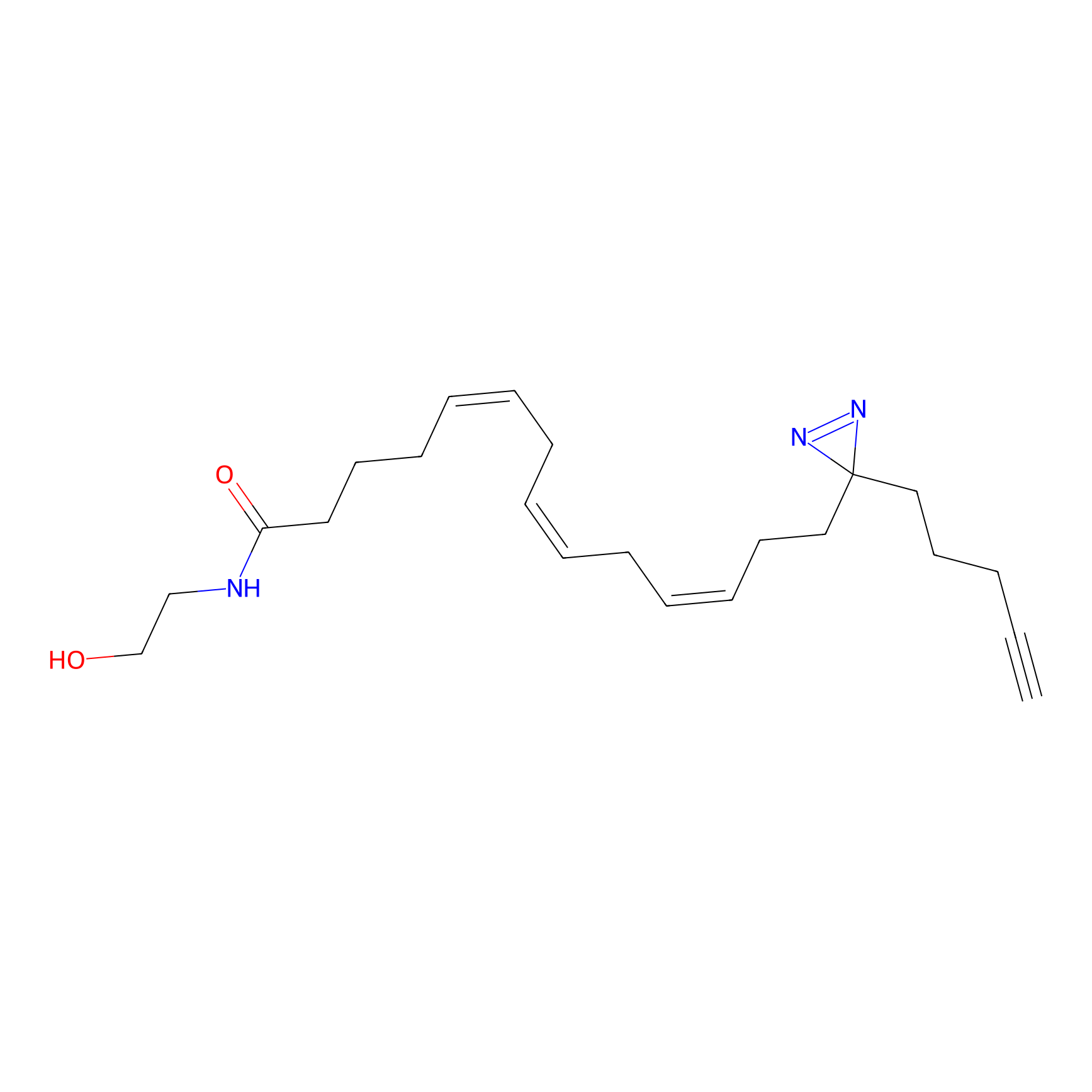 |
7.55 | LDD0146 | [12] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C133(0.66); C186(0.85) | LDD2142 | [6] |
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C370(0.77); C186(0.86) | LDD2112 | [6] |
| LDCM0502 | 1-(Cyanoacetyl)piperidine | MDA-MB-231 | C370(0.60); C133(0.48) | LDD2095 | [6] |
| LDCM0537 | 2-Cyano-N,N-dimethylacetamide | MDA-MB-231 | C370(0.85) | LDD2130 | [6] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C370(0.54); C133(0.83); C186(0.55); C126(0.75) | LDD2117 | [6] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C370(1.34); C186(0.92) | LDD2152 | [6] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C370(1.03); C186(0.66) | LDD2103 | [6] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C370(0.42) | LDD2132 | [6] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C370(0.57); C133(0.70); C186(0.76) | LDD2113 | [6] |
| LDCM0151 | AZ-11 | HeLa | 10.00 | LDD2154 | [5] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C133(0.85) | LDD2091 | [6] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C186(6.99); C133(2.96); C370(2.74) | LDD1702 | [6] |
| LDCM0082 | FK866 | A-549 | 7.55 | LDD0146 | [12] |
| LDCM0022 | KB02 | 697 | C370(1.65); C126(1.34); C133(1.64) | LDD2245 | [4] |
| LDCM0023 | KB03 | MDA-MB-231 | C186(14.14); C370(8.82); C133(4.42) | LDD1701 | [6] |
| LDCM0024 | KB05 | HMCB | C370(1.71) | LDD3312 | [4] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C370(0.99); C186(0.82) | LDD2102 | [6] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C370(0.65) | LDD2121 | [6] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C133(1.04) | LDD2089 | [6] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C370(1.54); C186(1.12) | LDD2090 | [6] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C370(1.07); C133(1.04); C186(1.13) | LDD2092 | [6] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C370(1.17); C186(0.54) | LDD2093 | [6] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C370(1.14); C133(1.30); C186(0.77) | LDD2094 | [6] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C370(0.71); C186(0.90) | LDD2097 | [6] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C370(1.09); C133(1.03); C186(0.89) | LDD2098 | [6] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C370(1.04); C133(1.41); C186(0.71) | LDD2099 | [6] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C370(0.58); C133(0.55); C186(0.80); C50(0.09) | LDD2100 | [6] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C370(0.65); C133(0.68) | LDD2101 | [6] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C370(0.66); C186(0.65) | LDD2104 | [6] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C370(1.37); C133(1.05); C186(0.85) | LDD2105 | [6] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C186(0.96) | LDD2106 | [6] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C370(0.96); C133(1.26); C186(0.74); C50(1.28) | LDD2107 | [6] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C370(0.96); C133(0.85); C186(0.74) | LDD2108 | [6] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C370(0.78); C133(0.67); C186(0.85); C126(0.82) | LDD2109 | [6] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C370(1.21); C133(1.27); C186(0.86) | LDD2111 | [6] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C370(0.61); C186(0.86) | LDD2114 | [6] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C370(0.44); C133(0.43); C186(0.53) | LDD2115 | [6] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C186(0.90) | LDD2118 | [6] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C370(2.18); C133(0.81); C186(1.45) | LDD2119 | [6] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C370(1.03) | LDD2120 | [6] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C370(0.56); C186(0.86) | LDD2122 | [6] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C370(0.98); C133(0.99); C186(0.88); C50(1.25) | LDD2123 | [6] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C370(0.14); C133(0.31) | LDD2124 | [6] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C370(0.82); C133(1.02); C186(0.59); C126(0.97) | LDD2125 | [6] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C370(0.12); C133(0.37); C186(1.02) | LDD2126 | [6] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C370(0.60); C133(0.83); C186(0.53); C50(1.05) | LDD2127 | [6] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C370(1.19); C133(1.04) | LDD2129 | [6] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C370(0.50) | LDD2133 | [6] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C370(0.46) | LDD2134 | [6] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C370(1.43); C186(1.09); C50(1.37) | LDD2135 | [6] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C370(1.32); C133(1.66); C186(0.81); C126(1.31) | LDD2136 | [6] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C370(1.01); C133(1.07); C186(0.81); C126(1.06) | LDD2137 | [6] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C370(2.25); C186(1.55) | LDD1700 | [6] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C370(0.87); C133(1.22); C186(0.52) | LDD2140 | [6] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C370(0.56); C186(0.57) | LDD2141 | [6] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C370(3.34); C133(1.18); C186(1.85) | LDD2144 | [6] |
| LDCM0551 | Nucleophilic fragment 5b | MDA-MB-231 | C370(0.92); C186(0.70) | LDD2145 | [6] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C370(0.97); C133(1.21); C186(0.67) | LDD2146 | [6] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C370(0.49); C133(0.39); C186(0.42) | LDD2148 | [6] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C370(0.63) | LDD2150 | [6] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C186(1.44) | LDD2153 | [6] |
| LDCM0112 | W16 | Hep-G2 | K398(1.38) | LDD0239 | [2] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Dopamine beta-hydroxylase (DBH) | Copper type II ascorbate-dependent monooxygenase family | P09172 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Junctophilin-3 (JPH3) | Junctophilin family | Q8WXH2 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Glial fibrillary acidic protein (GFAP) | Intermediate filament family | P14136 | |||
References
