Details of the Target
General Information of Target
| Target ID | LDTP06284 | |||||
|---|---|---|---|---|---|---|
| Target Name | TNFAIP3-interacting protein 1 (TNIP1) | |||||
| Gene Name | TNIP1 | |||||
| Gene ID | 10318 | |||||
| Synonyms |
KIAA0113; NAF1; TNFAIP3-interacting protein 1; A20-binding inhibitor of NF-kappa-B activation 1; ABIN-1; HIV-1 Nef-interacting protein; Nef-associated factor 1; Naf1; Nip40-1; Virion-associated nuclear shuttling protein; VAN; hVAN
|
|||||
| 3D Structure | ||||||
| Sequence |
MEGRGPYRIYDPGGSVPSGEASAAFERLVKENSRLKEKMQGIKMLGELLEESQMEATRLR
QKAEELVKDNELLPPPSPSLGSFDPLAELTGKDSNVTASPTAPACPSDKPAPVQKPPSSG TSSEFEVVTPEEQNSPESSSHANAMALGPLPREDGNLMLHLQRLETTLSVCAEEPDHGQL FTHLGRMALEFNRLASKVHKNEQRTSILQTLCEQLRKENEALKAKLDKGLEQRDQAAERL REENLELKKLLMSNGNKEGASGRPGSPKMEGTGKKAVAGQQQASVTAGKVPEVVALGAAE KKVKMLEQQRSELLEVNKQWDQHFRSMKQQYEQKITELRQKLADLQKQVTDLEAEREQKQ RDFDRKLLLAKSKIEMEETDKEQLTAEAKELRQKVKYLQDQLSPLTRQREYQEKEIQRLN KALEEALSIQTPPSSPPTAFGSPEGAGALLRKQELVTQNELLKQQVKIFEEDFQRERSDR ERMNEEKEELKKQVEKLQAQVTLSNAQLKAFKDEEKAREALRQQKRKAKASGERYHVEPH PEHLCGAYPYAYPPMPAMVPHHGFEDWSQIRYPPPPMAMEHPPPLPNSRLFHLPEYTWRL PCGGVRNPNQSSQVMDPPTARPTEPESPKNDREGPQ |
|||||
| Target Bioclass |
Other
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Inhibits NF-kappa-B activation and TNF-induced NF-kappa-B-dependent gene expression by regulating TAX1BP1 and A20/TNFAIP3-mediated deubiquitination of IKBKG; proposed to link A20/TNFAIP3 to ubiquitinated IKBKG. Involved in regulation of EGF-induced ERK1/ERK2 signaling pathway; blocks MAPK3/MAPK1 nuclear translocation and MAPK1-dependent transcription. Increases cell surface CD4(T4) antigen expression. Involved in the anti-inflammatory response of macrophages and positively regulates TLR-induced activation of CEBPB. Involved in the prevention of autoimmunity; this function implicates binding to polyubiquitin. Involved in leukocyte integrin activation during inflammation; this function is mediated by association with SELPLG and dependent on phosphorylation by SRC-family kinases. Interacts with HIV-1 matrix protein and is packaged into virions and overexpression can inhibit viral replication. May regulate matrix nuclear localization, both nuclear import of PIC (Preintegration complex) and export of GAG polyprotein and viral genomic RNA during virion production. In case of infection, promotes association of IKBKG with Shigella flexneri E3 ubiquitin-protein ligase ipah9.8 p which in turn promotes polyubiquitination of IKBKG leading to its proteasome-dependent degradation and thus is perturbing NF-kappa-B activation during bacterial infection.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
STPyne Probe Info |
 |
K200(5.96); K334(1.66); K452(5.73) | LDD0277 | [2] | |
|
BTD Probe Info |
 |
C602(2.23) | LDD2141 | [3] | |
|
NAIA_5 Probe Info |
 |
C212(20.00) | LDD2227 | [4] | |
|
HPAP Probe Info |
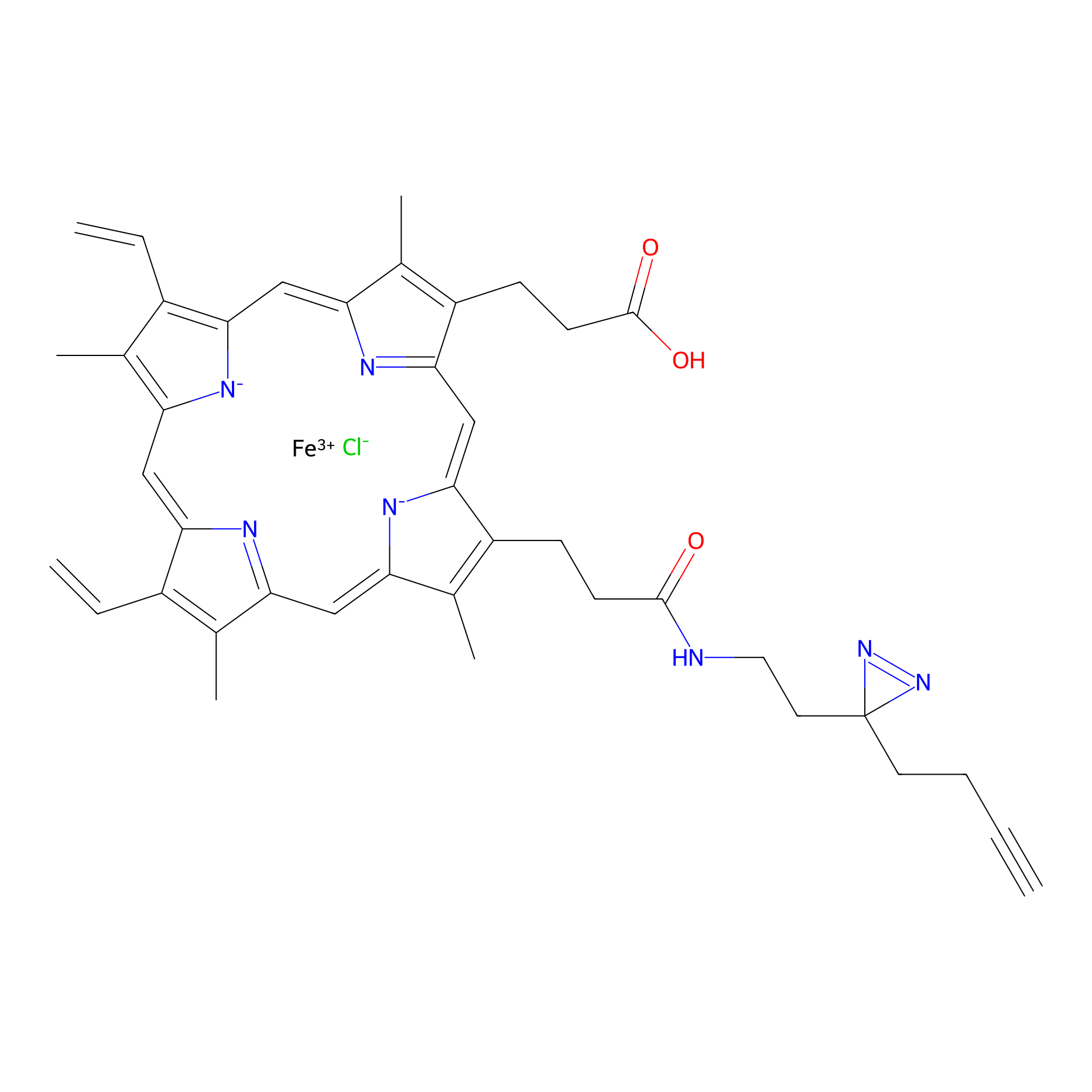 |
5.28 | LDD0062 | [5] | |
|
DBIA Probe Info |
 |
C212(0.95) | LDD0531 | [6] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [7] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [4] | |
|
IPM Probe Info |
 |
N.A. | LDD0147 | [8] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [8] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [9] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HCT 116 | C212(0.95) | LDD0531 | [6] |
| LDCM0215 | AC10 | HCT 116 | C212(1.07) | LDD0532 | [6] |
| LDCM0216 | AC100 | HCT 116 | C212(1.05) | LDD0533 | [6] |
| LDCM0217 | AC101 | HCT 116 | C212(1.02) | LDD0534 | [6] |
| LDCM0218 | AC102 | HCT 116 | C212(0.89) | LDD0535 | [6] |
| LDCM0219 | AC103 | HCT 116 | C212(1.00) | LDD0536 | [6] |
| LDCM0220 | AC104 | HCT 116 | C212(0.80) | LDD0537 | [6] |
| LDCM0221 | AC105 | HCT 116 | C212(1.05) | LDD0538 | [6] |
| LDCM0222 | AC106 | HCT 116 | C212(1.05) | LDD0539 | [6] |
| LDCM0223 | AC107 | HCT 116 | C212(0.99) | LDD0540 | [6] |
| LDCM0224 | AC108 | HCT 116 | C212(0.98) | LDD0541 | [6] |
| LDCM0225 | AC109 | HCT 116 | C212(1.02) | LDD0542 | [6] |
| LDCM0226 | AC11 | HCT 116 | C212(1.13) | LDD0543 | [6] |
| LDCM0227 | AC110 | HCT 116 | C212(1.28) | LDD0544 | [6] |
| LDCM0228 | AC111 | HCT 116 | C212(1.05) | LDD0545 | [6] |
| LDCM0229 | AC112 | HCT 116 | C212(1.10) | LDD0546 | [6] |
| LDCM0230 | AC113 | HCT 116 | C212(1.08) | LDD0547 | [6] |
| LDCM0231 | AC114 | HCT 116 | C212(0.98) | LDD0548 | [6] |
| LDCM0232 | AC115 | HCT 116 | C212(0.88) | LDD0549 | [6] |
| LDCM0233 | AC116 | HCT 116 | C212(1.04) | LDD0550 | [6] |
| LDCM0234 | AC117 | HCT 116 | C212(0.91) | LDD0551 | [6] |
| LDCM0235 | AC118 | HCT 116 | C212(0.95) | LDD0552 | [6] |
| LDCM0236 | AC119 | HCT 116 | C212(1.14) | LDD0553 | [6] |
| LDCM0237 | AC12 | HCT 116 | C212(1.03) | LDD0554 | [6] |
| LDCM0238 | AC120 | HCT 116 | C212(1.03) | LDD0555 | [6] |
| LDCM0239 | AC121 | HCT 116 | C212(0.92) | LDD0556 | [6] |
| LDCM0240 | AC122 | HCT 116 | C212(1.24) | LDD0557 | [6] |
| LDCM0241 | AC123 | HCT 116 | C212(0.79) | LDD0558 | [6] |
| LDCM0242 | AC124 | HCT 116 | C212(1.00) | LDD0559 | [6] |
| LDCM0243 | AC125 | HCT 116 | C212(0.96) | LDD0560 | [6] |
| LDCM0244 | AC126 | HCT 116 | C212(0.98) | LDD0561 | [6] |
| LDCM0245 | AC127 | HCT 116 | C212(1.20) | LDD0562 | [6] |
| LDCM0246 | AC128 | HCT 116 | C212(1.02) | LDD0563 | [6] |
| LDCM0247 | AC129 | HCT 116 | C212(1.19) | LDD0564 | [6] |
| LDCM0249 | AC130 | HCT 116 | C212(1.07) | LDD0566 | [6] |
| LDCM0250 | AC131 | HCT 116 | C212(1.22) | LDD0567 | [6] |
| LDCM0251 | AC132 | HCT 116 | C212(1.25) | LDD0568 | [6] |
| LDCM0252 | AC133 | HCT 116 | C212(1.34) | LDD0569 | [6] |
| LDCM0253 | AC134 | HCT 116 | C212(1.09) | LDD0570 | [6] |
| LDCM0254 | AC135 | HCT 116 | C212(0.93) | LDD0571 | [6] |
| LDCM0255 | AC136 | HCT 116 | C212(1.51) | LDD0572 | [6] |
| LDCM0256 | AC137 | HCT 116 | C212(1.15) | LDD0573 | [6] |
| LDCM0257 | AC138 | HCT 116 | C212(1.01) | LDD0574 | [6] |
| LDCM0258 | AC139 | HCT 116 | C212(1.28) | LDD0575 | [6] |
| LDCM0259 | AC14 | HCT 116 | C212(1.00) | LDD0576 | [6] |
| LDCM0260 | AC140 | HCT 116 | C212(1.22) | LDD0577 | [6] |
| LDCM0261 | AC141 | HCT 116 | C212(1.24) | LDD0578 | [6] |
| LDCM0262 | AC142 | HCT 116 | C212(1.19) | LDD0579 | [6] |
| LDCM0270 | AC15 | HCT 116 | C212(1.04) | LDD0587 | [6] |
| LDCM0276 | AC17 | HCT 116 | C212(0.97) | LDD0593 | [6] |
| LDCM0277 | AC18 | HCT 116 | C212(0.90) | LDD0594 | [6] |
| LDCM0278 | AC19 | HCT 116 | C212(1.00) | LDD0595 | [6] |
| LDCM0279 | AC2 | HCT 116 | C212(1.00) | LDD0596 | [6] |
| LDCM0280 | AC20 | HCT 116 | C212(1.03) | LDD0597 | [6] |
| LDCM0281 | AC21 | HCT 116 | C212(1.05) | LDD0598 | [6] |
| LDCM0282 | AC22 | HCT 116 | C212(0.98) | LDD0599 | [6] |
| LDCM0283 | AC23 | HCT 116 | C212(1.01) | LDD0600 | [6] |
| LDCM0284 | AC24 | HCT 116 | C212(1.04) | LDD0601 | [6] |
| LDCM0285 | AC25 | HCT 116 | C212(0.98) | LDD0602 | [6] |
| LDCM0286 | AC26 | HCT 116 | C212(1.17) | LDD0603 | [6] |
| LDCM0287 | AC27 | HCT 116 | C212(1.04) | LDD0604 | [6] |
| LDCM0288 | AC28 | HCT 116 | C212(1.12) | LDD0605 | [6] |
| LDCM0289 | AC29 | HCT 116 | C212(1.03) | LDD0606 | [6] |
| LDCM0290 | AC3 | HCT 116 | C212(1.02) | LDD0607 | [6] |
| LDCM0291 | AC30 | HCT 116 | C212(1.19) | LDD0608 | [6] |
| LDCM0292 | AC31 | HCT 116 | C212(1.07) | LDD0609 | [6] |
| LDCM0293 | AC32 | HCT 116 | C212(1.19) | LDD0610 | [6] |
| LDCM0294 | AC33 | HCT 116 | C212(1.05) | LDD0611 | [6] |
| LDCM0295 | AC34 | HCT 116 | C212(0.96) | LDD0612 | [6] |
| LDCM0296 | AC35 | HEK-293T | C212(0.82) | LDD0894 | [6] |
| LDCM0297 | AC36 | HEK-293T | C212(1.46) | LDD0895 | [6] |
| LDCM0298 | AC37 | HEK-293T | C212(1.85) | LDD0896 | [6] |
| LDCM0299 | AC38 | HEK-293T | C212(1.70) | LDD0897 | [6] |
| LDCM0300 | AC39 | HEK-293T | C212(1.12) | LDD0898 | [6] |
| LDCM0301 | AC4 | HCT 116 | C212(1.01) | LDD0618 | [6] |
| LDCM0302 | AC40 | HEK-293T | C212(1.07) | LDD0900 | [6] |
| LDCM0303 | AC41 | HEK-293T | C212(1.05) | LDD0901 | [6] |
| LDCM0304 | AC42 | HEK-293T | C212(1.17) | LDD0902 | [6] |
| LDCM0305 | AC43 | HEK-293T | C212(0.96) | LDD0903 | [6] |
| LDCM0306 | AC44 | HEK-293T | C212(1.33) | LDD0904 | [6] |
| LDCM0307 | AC45 | HEK-293T | C212(1.73) | LDD0905 | [6] |
| LDCM0308 | AC46 | HEK-293T | C212(0.83) | LDD0906 | [6] |
| LDCM0309 | AC47 | HEK-293T | C212(0.98) | LDD0907 | [6] |
| LDCM0310 | AC48 | HEK-293T | C212(1.30) | LDD0908 | [6] |
| LDCM0311 | AC49 | HEK-293T | C212(1.30) | LDD0909 | [6] |
| LDCM0312 | AC5 | HCT 116 | C212(1.06) | LDD0629 | [6] |
| LDCM0313 | AC50 | HEK-293T | C212(1.11) | LDD0911 | [6] |
| LDCM0314 | AC51 | HEK-293T | C212(1.73) | LDD0912 | [6] |
| LDCM0315 | AC52 | HEK-293T | C212(1.55) | LDD0913 | [6] |
| LDCM0316 | AC53 | HEK-293T | C212(1.39) | LDD0914 | [6] |
| LDCM0317 | AC54 | HEK-293T | C212(1.38) | LDD0915 | [6] |
| LDCM0318 | AC55 | HEK-293T | C212(1.43) | LDD0916 | [6] |
| LDCM0319 | AC56 | HEK-293T | C212(1.25) | LDD0917 | [6] |
| LDCM0320 | AC57 | HCT 116 | C212(0.92) | LDD0637 | [6] |
| LDCM0321 | AC58 | HCT 116 | C212(0.99) | LDD0638 | [6] |
| LDCM0322 | AC59 | HCT 116 | C212(0.91) | LDD0639 | [6] |
| LDCM0323 | AC6 | HCT 116 | C212(1.35) | LDD0640 | [6] |
| LDCM0324 | AC60 | HCT 116 | C212(1.07) | LDD0641 | [6] |
| LDCM0325 | AC61 | HCT 116 | C212(1.02) | LDD0642 | [6] |
| LDCM0326 | AC62 | HCT 116 | C212(0.86) | LDD0643 | [6] |
| LDCM0327 | AC63 | HCT 116 | C212(1.04) | LDD0644 | [6] |
| LDCM0328 | AC64 | HCT 116 | C212(1.34) | LDD0645 | [6] |
| LDCM0329 | AC65 | HCT 116 | C212(1.01) | LDD0646 | [6] |
| LDCM0330 | AC66 | HCT 116 | C212(0.92) | LDD0647 | [6] |
| LDCM0331 | AC67 | HCT 116 | C212(0.95) | LDD0648 | [6] |
| LDCM0332 | AC68 | HEK-293T | C212(0.98) | LDD0930 | [6] |
| LDCM0333 | AC69 | HEK-293T | C212(0.88) | LDD0931 | [6] |
| LDCM0334 | AC7 | HCT 116 | C212(1.06) | LDD0651 | [6] |
| LDCM0335 | AC70 | HEK-293T | C212(1.07) | LDD0933 | [6] |
| LDCM0336 | AC71 | HEK-293T | C212(0.88) | LDD0934 | [6] |
| LDCM0337 | AC72 | HEK-293T | C212(1.03) | LDD0935 | [6] |
| LDCM0338 | AC73 | HEK-293T | C212(0.92) | LDD0936 | [6] |
| LDCM0339 | AC74 | HEK-293T | C212(0.88) | LDD0937 | [6] |
| LDCM0340 | AC75 | HEK-293T | C212(1.18) | LDD0938 | [6] |
| LDCM0341 | AC76 | HEK-293T | C212(1.15) | LDD0939 | [6] |
| LDCM0342 | AC77 | HEK-293T | C212(0.94) | LDD0940 | [6] |
| LDCM0343 | AC78 | HEK-293T | C212(1.07) | LDD0941 | [6] |
| LDCM0344 | AC79 | HEK-293T | C212(0.87) | LDD0942 | [6] |
| LDCM0345 | AC8 | HCT 116 | C212(0.91) | LDD0662 | [6] |
| LDCM0346 | AC80 | HEK-293T | C212(1.00) | LDD0944 | [6] |
| LDCM0347 | AC81 | HEK-293T | C212(0.92) | LDD0945 | [6] |
| LDCM0348 | AC82 | HEK-293T | C212(1.30) | LDD0946 | [6] |
| LDCM0349 | AC83 | HEK-293T | C212(1.15) | LDD0947 | [6] |
| LDCM0350 | AC84 | HEK-293T | C212(1.69) | LDD0948 | [6] |
| LDCM0351 | AC85 | HEK-293T | C212(1.16) | LDD0949 | [6] |
| LDCM0352 | AC86 | HEK-293T | C212(1.35) | LDD0950 | [6] |
| LDCM0353 | AC87 | HEK-293T | C212(1.33) | LDD0951 | [6] |
| LDCM0354 | AC88 | HEK-293T | C212(1.55) | LDD0952 | [6] |
| LDCM0355 | AC89 | HEK-293T | C212(1.45) | LDD0953 | [6] |
| LDCM0357 | AC90 | HEK-293T | C212(1.46) | LDD0955 | [6] |
| LDCM0358 | AC91 | HEK-293T | C212(1.32) | LDD0956 | [6] |
| LDCM0359 | AC92 | HEK-293T | C212(1.55) | LDD0957 | [6] |
| LDCM0360 | AC93 | HEK-293T | C212(1.57) | LDD0958 | [6] |
| LDCM0361 | AC94 | HEK-293T | C212(1.61) | LDD0959 | [6] |
| LDCM0362 | AC95 | HEK-293T | C212(1.44) | LDD0960 | [6] |
| LDCM0363 | AC96 | HEK-293T | C212(1.54) | LDD0961 | [6] |
| LDCM0364 | AC97 | HEK-293T | C212(2.59) | LDD0962 | [6] |
| LDCM0365 | AC98 | HCT 116 | C212(1.22) | LDD0682 | [6] |
| LDCM0366 | AC99 | HCT 116 | C212(1.16) | LDD0683 | [6] |
| LDCM0248 | AKOS034007472 | HCT 116 | C212(1.13) | LDD0565 | [6] |
| LDCM0356 | AKOS034007680 | HCT 116 | C212(1.08) | LDD0673 | [6] |
| LDCM0275 | AKOS034007705 | HCT 116 | C212(1.45) | LDD0592 | [6] |
| LDCM0156 | Aniline | NCI-H1299 | 12.92 | LDD0403 | [1] |
| LDCM0632 | CL-Sc | Hep-G2 | C212(20.00) | LDD2227 | [4] |
| LDCM0367 | CL1 | HEK-293T | C602(1.57); C105(0.79); C171(0.98) | LDD1571 | [10] |
| LDCM0368 | CL10 | HEK-293T | C602(2.52); C105(1.11); C171(1.18) | LDD1572 | [10] |
| LDCM0369 | CL100 | HCT 116 | C212(0.99) | LDD0686 | [6] |
| LDCM0370 | CL101 | HCT 116 | C212(1.20) | LDD0687 | [6] |
| LDCM0371 | CL102 | HCT 116 | C212(1.06) | LDD0688 | [6] |
| LDCM0372 | CL103 | HCT 116 | C212(0.97) | LDD0689 | [6] |
| LDCM0373 | CL104 | HCT 116 | C212(1.08) | LDD0690 | [6] |
| LDCM0374 | CL105 | HCT 116 | C212(1.02) | LDD0691 | [6] |
| LDCM0375 | CL106 | HCT 116 | C212(1.17) | LDD0692 | [6] |
| LDCM0376 | CL107 | HCT 116 | C212(1.16) | LDD0693 | [6] |
| LDCM0377 | CL108 | HCT 116 | C212(1.09) | LDD0694 | [6] |
| LDCM0378 | CL109 | HCT 116 | C212(1.02) | LDD0695 | [6] |
| LDCM0379 | CL11 | HEK-293T | C602(1.69); C105(1.80); C171(0.94) | LDD1583 | [10] |
| LDCM0380 | CL110 | HCT 116 | C212(0.99) | LDD0697 | [6] |
| LDCM0381 | CL111 | HCT 116 | C212(0.94) | LDD0698 | [6] |
| LDCM0382 | CL112 | HCT 116 | C212(1.11) | LDD0699 | [6] |
| LDCM0383 | CL113 | HCT 116 | C212(1.00) | LDD0700 | [6] |
| LDCM0384 | CL114 | HCT 116 | C212(1.10) | LDD0701 | [6] |
| LDCM0385 | CL115 | HCT 116 | C212(1.12) | LDD0702 | [6] |
| LDCM0386 | CL116 | HCT 116 | C212(1.02) | LDD0703 | [6] |
| LDCM0387 | CL117 | HEK-293T | C212(1.29) | LDD0985 | [6] |
| LDCM0388 | CL118 | HEK-293T | C212(1.27) | LDD0986 | [6] |
| LDCM0389 | CL119 | HEK-293T | C212(1.09) | LDD0987 | [6] |
| LDCM0390 | CL12 | HEK-293T | C171(1.45) | LDD1594 | [10] |
| LDCM0391 | CL120 | HEK-293T | C212(1.34) | LDD0989 | [6] |
| LDCM0392 | CL121 | HEK-293T | C212(1.64) | LDD0990 | [6] |
| LDCM0393 | CL122 | HEK-293T | C212(0.89) | LDD0991 | [6] |
| LDCM0394 | CL123 | HEK-293T | C212(0.95) | LDD0992 | [6] |
| LDCM0395 | CL124 | HEK-293T | C212(1.25) | LDD0993 | [6] |
| LDCM0396 | CL125 | HCT 116 | C212(1.02) | LDD0713 | [6] |
| LDCM0397 | CL126 | HCT 116 | C212(0.88) | LDD0714 | [6] |
| LDCM0398 | CL127 | HCT 116 | C212(0.90) | LDD0715 | [6] |
| LDCM0399 | CL128 | HCT 116 | C212(0.99) | LDD0716 | [6] |
| LDCM0400 | CL13 | HEK-293T | C602(1.29); C105(1.16); C171(1.17) | LDD1604 | [10] |
| LDCM0401 | CL14 | HEK-293T | C602(1.19); C105(0.94) | LDD1605 | [10] |
| LDCM0402 | CL15 | HEK-293T | C602(1.32); C105(0.86) | LDD1606 | [10] |
| LDCM0403 | CL16 | HCT 116 | C212(1.24) | LDD0720 | [6] |
| LDCM0404 | CL17 | HEK-293T | C171(0.97) | LDD1608 | [10] |
| LDCM0406 | CL19 | HEK-293T | C171(1.03) | LDD1610 | [10] |
| LDCM0407 | CL2 | HEK-293T | C602(1.13); C105(1.02) | LDD1611 | [10] |
| LDCM0408 | CL20 | HEK-293T | C171(1.05) | LDD1612 | [10] |
| LDCM0410 | CL22 | HEK-293T | C602(1.34); C105(1.69); C171(1.01) | LDD1614 | [10] |
| LDCM0411 | CL23 | HEK-293T | C602(1.16); C105(1.92); C171(1.01) | LDD1615 | [10] |
| LDCM0412 | CL24 | HEK-293T | C171(0.91) | LDD1616 | [10] |
| LDCM0413 | CL25 | HEK-293T | C602(0.96); C105(1.18); C171(1.21) | LDD1617 | [10] |
| LDCM0414 | CL26 | HEK-293T | C602(1.06); C105(1.03) | LDD1618 | [10] |
| LDCM0415 | CL27 | HEK-293T | C602(0.86); C105(0.83) | LDD1619 | [10] |
| LDCM0416 | CL28 | HEK-293T | C105(0.94) | LDD1620 | [10] |
| LDCM0417 | CL29 | HEK-293T | C171(0.94) | LDD1621 | [10] |
| LDCM0418 | CL3 | HEK-293T | C602(0.97); C105(1.02) | LDD1622 | [10] |
| LDCM0420 | CL31 | HCT 116 | C212(0.94) | LDD0737 | [6] |
| LDCM0421 | CL32 | HCT 116 | C212(1.22) | LDD0738 | [6] |
| LDCM0422 | CL33 | HCT 116 | C212(1.13) | LDD0739 | [6] |
| LDCM0423 | CL34 | HCT 116 | C212(1.15) | LDD0740 | [6] |
| LDCM0424 | CL35 | HCT 116 | C212(1.13) | LDD0741 | [6] |
| LDCM0425 | CL36 | HCT 116 | C212(0.93) | LDD0742 | [6] |
| LDCM0426 | CL37 | HCT 116 | C212(1.11) | LDD0743 | [6] |
| LDCM0428 | CL39 | HCT 116 | C212(1.08) | LDD0745 | [6] |
| LDCM0429 | CL4 | HEK-293T | C105(0.80) | LDD1633 | [10] |
| LDCM0430 | CL40 | HCT 116 | C212(1.21) | LDD0747 | [6] |
| LDCM0431 | CL41 | HCT 116 | C212(0.97) | LDD0748 | [6] |
| LDCM0432 | CL42 | HCT 116 | C212(1.14) | LDD0749 | [6] |
| LDCM0433 | CL43 | HCT 116 | C212(1.22) | LDD0750 | [6] |
| LDCM0434 | CL44 | HCT 116 | C212(1.09) | LDD0751 | [6] |
| LDCM0435 | CL45 | HCT 116 | C212(1.06) | LDD0752 | [6] |
| LDCM0436 | CL46 | HCT 116 | C212(1.48) | LDD0753 | [6] |
| LDCM0437 | CL47 | HCT 116 | C212(1.21) | LDD0754 | [6] |
| LDCM0438 | CL48 | HCT 116 | C212(1.25) | LDD0755 | [6] |
| LDCM0439 | CL49 | HCT 116 | C212(1.25) | LDD0756 | [6] |
| LDCM0440 | CL5 | HEK-293T | C171(1.18) | LDD1644 | [10] |
| LDCM0441 | CL50 | HCT 116 | C212(1.40) | LDD0758 | [6] |
| LDCM0442 | CL51 | HCT 116 | C212(1.18) | LDD0759 | [6] |
| LDCM0443 | CL52 | HCT 116 | C212(1.17) | LDD0760 | [6] |
| LDCM0444 | CL53 | HCT 116 | C212(1.20) | LDD0761 | [6] |
| LDCM0445 | CL54 | HCT 116 | C212(1.39) | LDD0762 | [6] |
| LDCM0446 | CL55 | HCT 116 | C212(1.21) | LDD0763 | [6] |
| LDCM0447 | CL56 | HCT 116 | C212(1.17) | LDD0764 | [6] |
| LDCM0448 | CL57 | HCT 116 | C212(1.41) | LDD0765 | [6] |
| LDCM0449 | CL58 | HCT 116 | C212(1.16) | LDD0766 | [6] |
| LDCM0450 | CL59 | HCT 116 | C212(1.04) | LDD0767 | [6] |
| LDCM0452 | CL60 | HCT 116 | C212(1.46) | LDD0769 | [6] |
| LDCM0453 | CL61 | HEK-293T | C212(0.91) | LDD1051 | [6] |
| LDCM0454 | CL62 | HEK-293T | C212(1.63) | LDD1052 | [6] |
| LDCM0455 | CL63 | HEK-293T | C212(1.36) | LDD1053 | [6] |
| LDCM0456 | CL64 | HEK-293T | C212(1.00) | LDD1054 | [6] |
| LDCM0457 | CL65 | HEK-293T | C212(0.81) | LDD1055 | [6] |
| LDCM0458 | CL66 | HEK-293T | C212(1.05) | LDD1056 | [6] |
| LDCM0459 | CL67 | HEK-293T | C212(0.99) | LDD1057 | [6] |
| LDCM0460 | CL68 | HEK-293T | C212(1.10) | LDD1058 | [6] |
| LDCM0461 | CL69 | HEK-293T | C212(1.06) | LDD1059 | [6] |
| LDCM0462 | CL7 | HEK-293T | C171(0.97) | LDD1665 | [10] |
| LDCM0463 | CL70 | HEK-293T | C212(0.90) | LDD1061 | [6] |
| LDCM0464 | CL71 | HEK-293T | C212(1.18) | LDD1062 | [6] |
| LDCM0465 | CL72 | HEK-293T | C212(1.24) | LDD1063 | [6] |
| LDCM0466 | CL73 | HEK-293T | C212(1.12) | LDD1064 | [6] |
| LDCM0467 | CL74 | HEK-293T | C212(1.89) | LDD1065 | [6] |
| LDCM0469 | CL76 | HEK-293T | C105(0.74) | LDD1672 | [10] |
| LDCM0470 | CL77 | HEK-293T | C171(0.95) | LDD1673 | [10] |
| LDCM0472 | CL79 | HEK-293T | C171(1.07) | LDD1675 | [10] |
| LDCM0473 | CL8 | HEK-293T | C171(1.16) | LDD1676 | [10] |
| LDCM0474 | CL80 | HEK-293T | C171(0.96) | LDD1677 | [10] |
| LDCM0476 | CL82 | HEK-293T | C602(1.11); C105(1.46); C171(1.01) | LDD1679 | [10] |
| LDCM0477 | CL83 | HEK-293T | C602(1.47); C105(1.49); C171(0.85) | LDD1680 | [10] |
| LDCM0478 | CL84 | HEK-293T | C171(1.25) | LDD1681 | [10] |
| LDCM0479 | CL85 | HEK-293T | C602(0.90); C105(0.99); C171(1.01) | LDD1682 | [10] |
| LDCM0480 | CL86 | HEK-293T | C602(0.90); C105(0.96) | LDD1683 | [10] |
| LDCM0481 | CL87 | HEK-293T | C602(1.08); C105(1.01) | LDD1684 | [10] |
| LDCM0482 | CL88 | HEK-293T | C105(0.70) | LDD1685 | [10] |
| LDCM0483 | CL89 | HEK-293T | C171(1.02) | LDD1686 | [10] |
| LDCM0486 | CL91 | HCT 116 | C212(1.06) | LDD0803 | [6] |
| LDCM0487 | CL92 | HCT 116 | C212(1.15) | LDD0804 | [6] |
| LDCM0488 | CL93 | HCT 116 | C212(1.11) | LDD0805 | [6] |
| LDCM0489 | CL94 | HCT 116 | C212(1.03) | LDD0806 | [6] |
| LDCM0490 | CL95 | HCT 116 | C212(1.15) | LDD0807 | [6] |
| LDCM0491 | CL96 | HCT 116 | C212(1.04) | LDD0808 | [6] |
| LDCM0492 | CL97 | HCT 116 | C212(1.02) | LDD0809 | [6] |
| LDCM0493 | CL98 | HCT 116 | C212(1.08) | LDD0810 | [6] |
| LDCM0494 | CL99 | HCT 116 | C212(1.03) | LDD0811 | [6] |
| LDCM0495 | E2913 | HEK-293T | C602(1.14); C105(1.05) | LDD1698 | [10] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C602(1.59) | LDD1702 | [3] |
| LDCM0468 | Fragment33 | HEK-293T | C212(1.38) | LDD1066 | [6] |
| LDCM0427 | Fragment51 | HCT 116 | C212(1.44) | LDD0744 | [6] |
| LDCM0022 | KB02 | HEK-293T | C105(0.97) | LDD1492 | [10] |
| LDCM0023 | KB03 | HEK-293T | C105(0.98) | LDD1497 | [10] |
| LDCM0024 | KB05 | MEWO | C602(1.93) | LDD3319 | [11] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C602(2.23) | LDD2141 | [3] |
| LDCM0014 | Panhematin | HEK-293T | 5.28 | LDD0062 | [5] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| AP-1 complex subunit mu-1 (AP1M1) | Adaptor complexes medium subunit family | Q9BXS5 | |||
| Hsp90 co-chaperone Cdc37 (CDC37) | CDC37 family | Q16543 | |||
| Optineurin (OPTN) | . | Q96CV9 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Nuclear factor NF-kappa-B p105 subunit (NFKB1) | . | P19838 | |||
| Zinc finger and BTB domain-containing protein 25 (ZBTB25) | . | P24278 | |||
Other
References
