Details of the Target
General Information of Target
| Target ID | LDTP05001 | |||||
|---|---|---|---|---|---|---|
| Target Name | Large ribosomal subunit protein uL23 (RPL23A) | |||||
| Gene Name | RPL23A | |||||
| Gene ID | 6147 | |||||
| Synonyms |
Large ribosomal subunit protein uL23; 60S ribosomal protein L23a |
|||||
| 3D Structure | ||||||
| Sequence |
MAPKAKKEAPAPPKAEAKAKALKAKKAVLKGVHSHKKKKIRTSPTFRRPKTLRLRRQPKY
PRKSAPRRNKLDHYAIIKFPLTTESAMKKIEDNNTLVFIVDVKANKHQIKQAVKKLYDID VAKVNTLIRPDGEKKAYVRLAPDYDALDVANKIGII |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Universal ribosomal protein uL23 family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Component of the large ribosomal subunit. The ribosome is a large ribonucleoprotein complex responsible for the synthesis of proteins in the cell. Binds a specific region on the 26S rRNA. May promote p53/TP53 degradation possibly through the stimulation of MDM2-mediated TP53 polyubiquitination.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
P1 Probe Info |
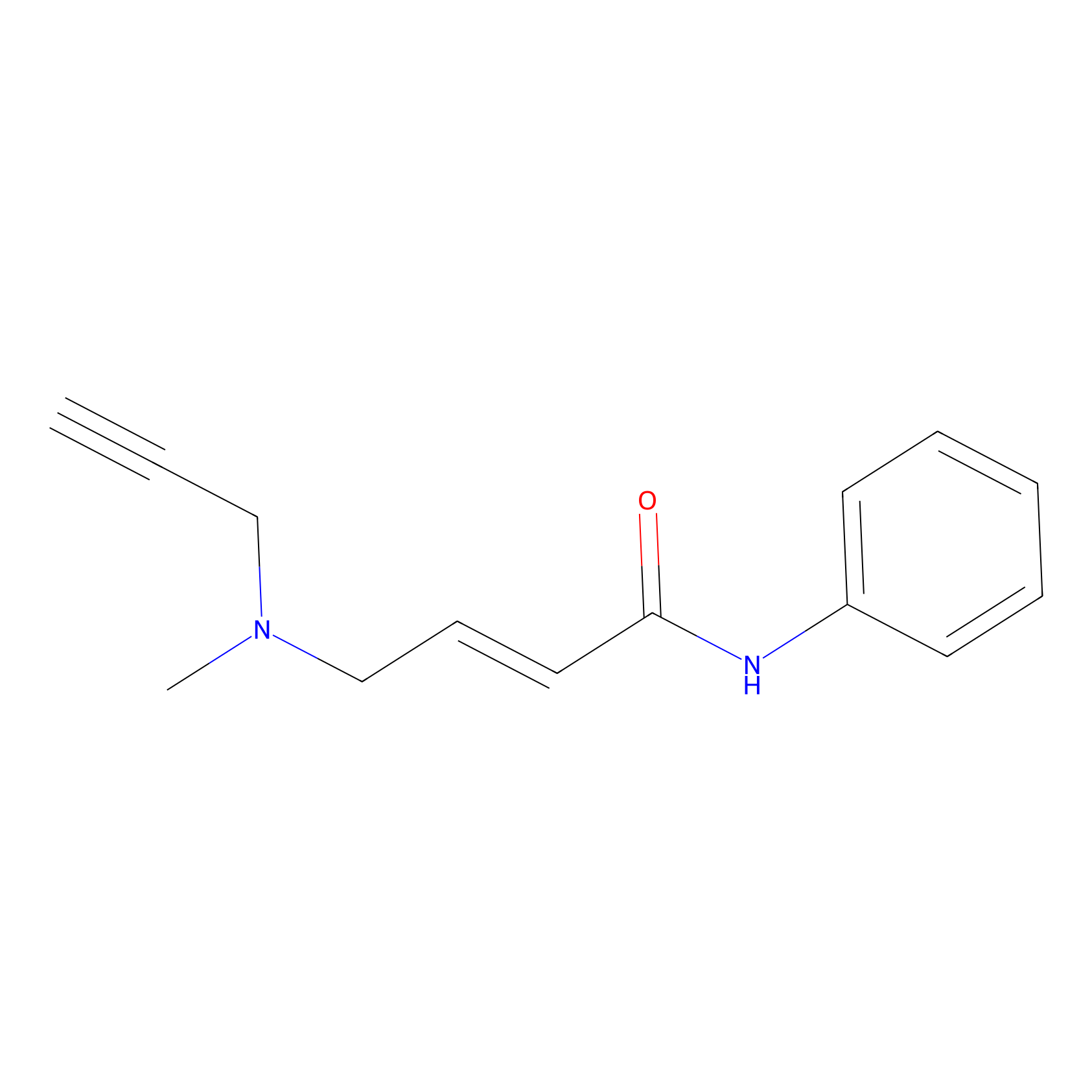 |
4.16 | LDD0448 | [1] | |
|
P2 Probe Info |
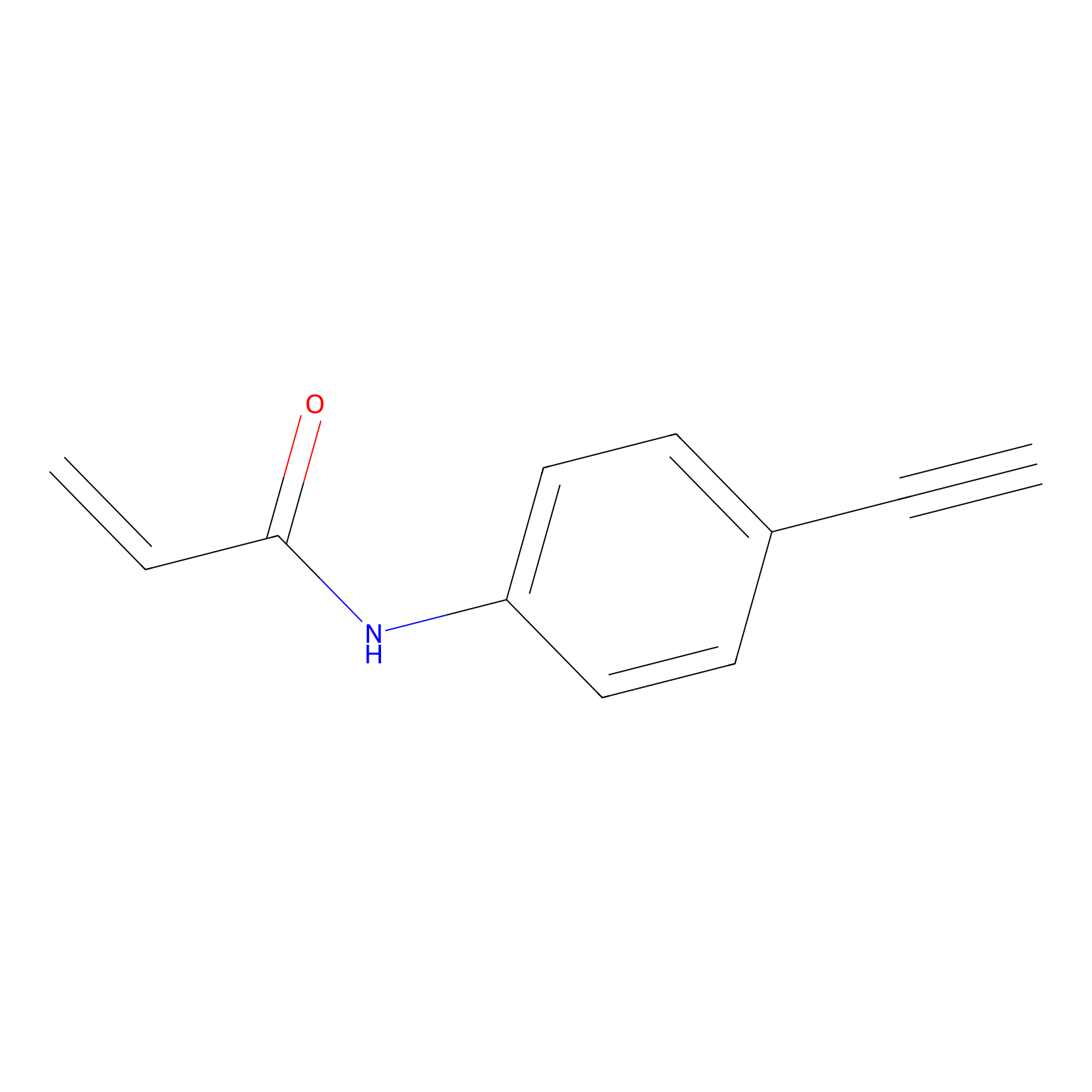 |
6.12 | LDD0449 | [1] | |
|
P3 Probe Info |
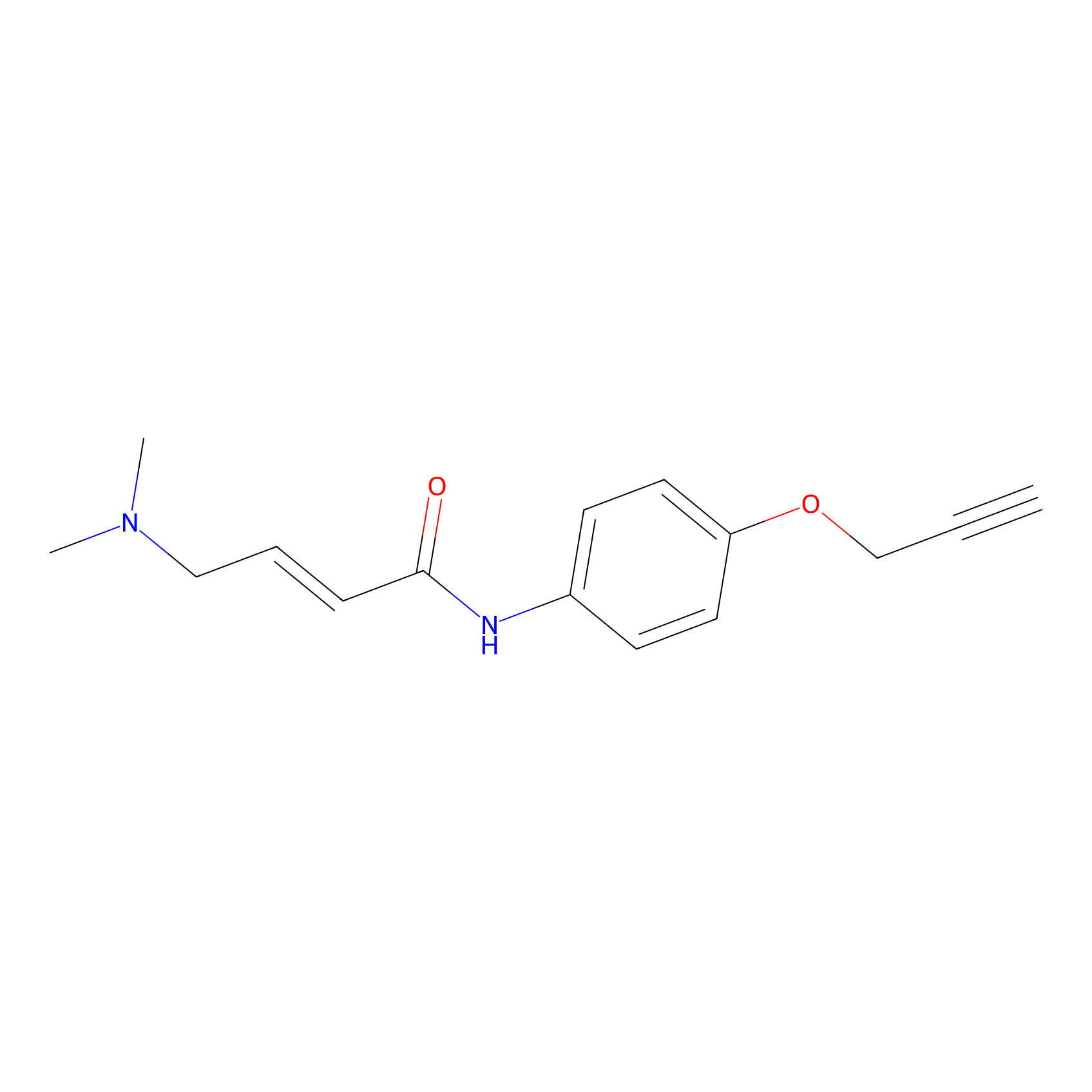 |
4.04 | LDD0450 | [1] | |
|
P8 Probe Info |
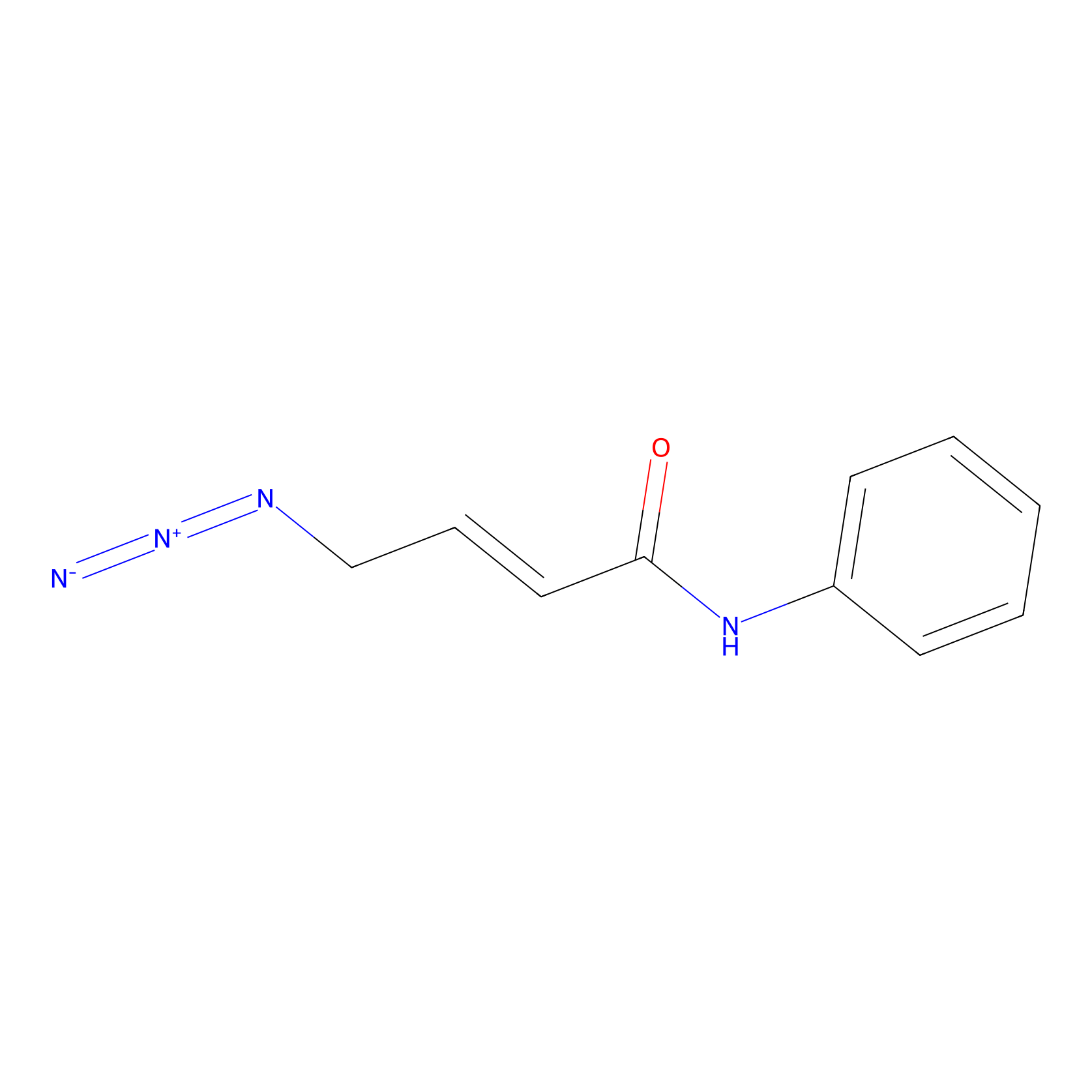 |
1.55 | LDD0451 | [1] | |
|
A-EBA Probe Info |
 |
2.58 | LDD0215 | [2] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [3] | |
|
C-Sul Probe Info |
 |
4.23 | LDD0066 | [4] | |
|
TH211 Probe Info |
 |
Y74(20.00) | LDD0257 | [5] | |
|
TH216 Probe Info |
 |
Y117(19.56); Y74(14.98); Y144(5.32) | LDD0259 | [5] | |
|
STPyne Probe Info |
 |
K106(9.09); K110(7.49); K115(4.98); K134(6.67) | LDD0277 | [6] | |
|
AZ-9 Probe Info |
 |
D143(1.07) | LDD2208 | [7] | |
|
ONAyne Probe Info |
 |
K115(0.00); K70(0.00) | LDD0273 | [6] | |
|
OPA-S-S-alkyne Probe Info |
 |
K152(4.64) | LDD3494 | [8] | |
|
Probe 1 Probe Info |
 |
Y74(24.24); Y144(111.35) | LDD3495 | [9] | |
|
HHS-465 Probe Info |
 |
Y74(10.00) | LDD2237 | [10] | |
|
AMP probe Probe Info |
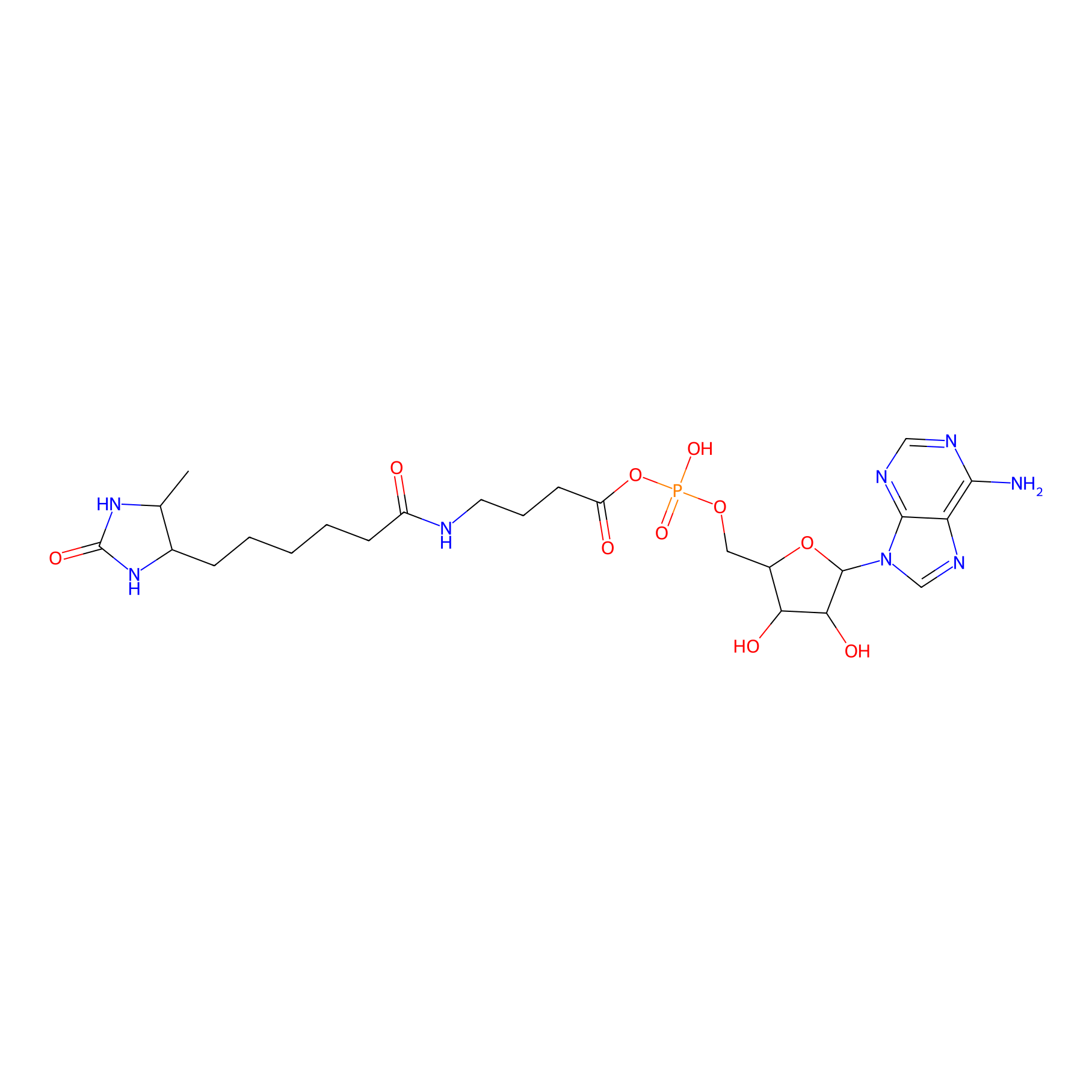 |
N.A. | LDD0200 | [11] | |
|
ATP probe Probe Info |
 |
K115(0.00); K123(0.00); K70(0.00); K134(0.00) | LDD0199 | [11] | |
|
m-APA Probe Info |
 |
N.A. | LDD2234 | [12] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [13] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [14] | |
|
SF Probe Info |
 |
K110(0.00); K7(0.00); K115(0.00); Y117(0.00) | LDD0028 | [15] | |
|
1c-yne Probe Info |
 |
K123(0.00); K78(0.00); K103(0.00); K89(0.00) | LDD0228 | [16] | |
|
1d-yne Probe Info |
 |
K70(0.00); K89(0.00) | LDD0357 | [16] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [17] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [17] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [17] | |
|
MPP-AC Probe Info |
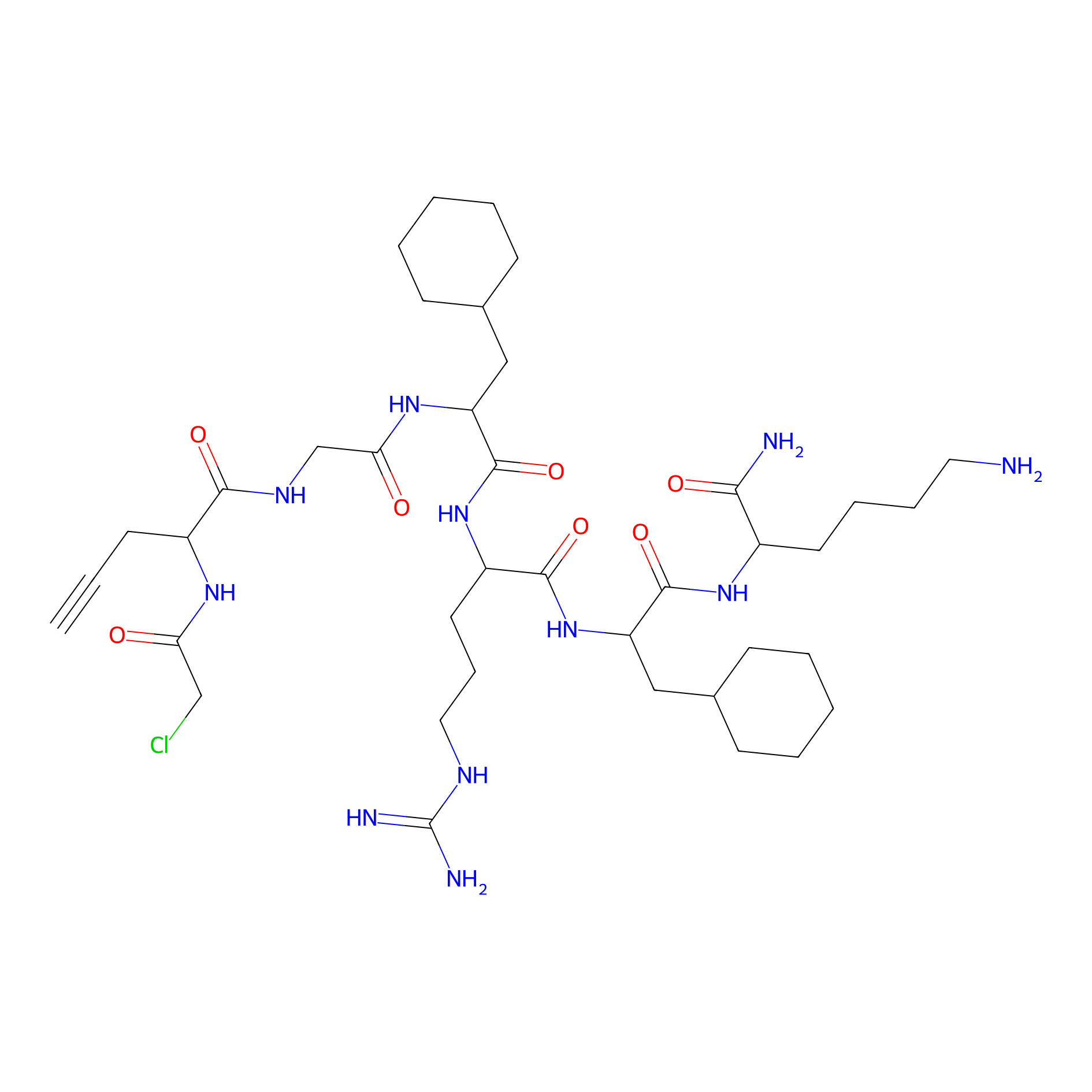 |
N.A. | LDD0428 | [18] | |
|
TPP-AC Probe Info |
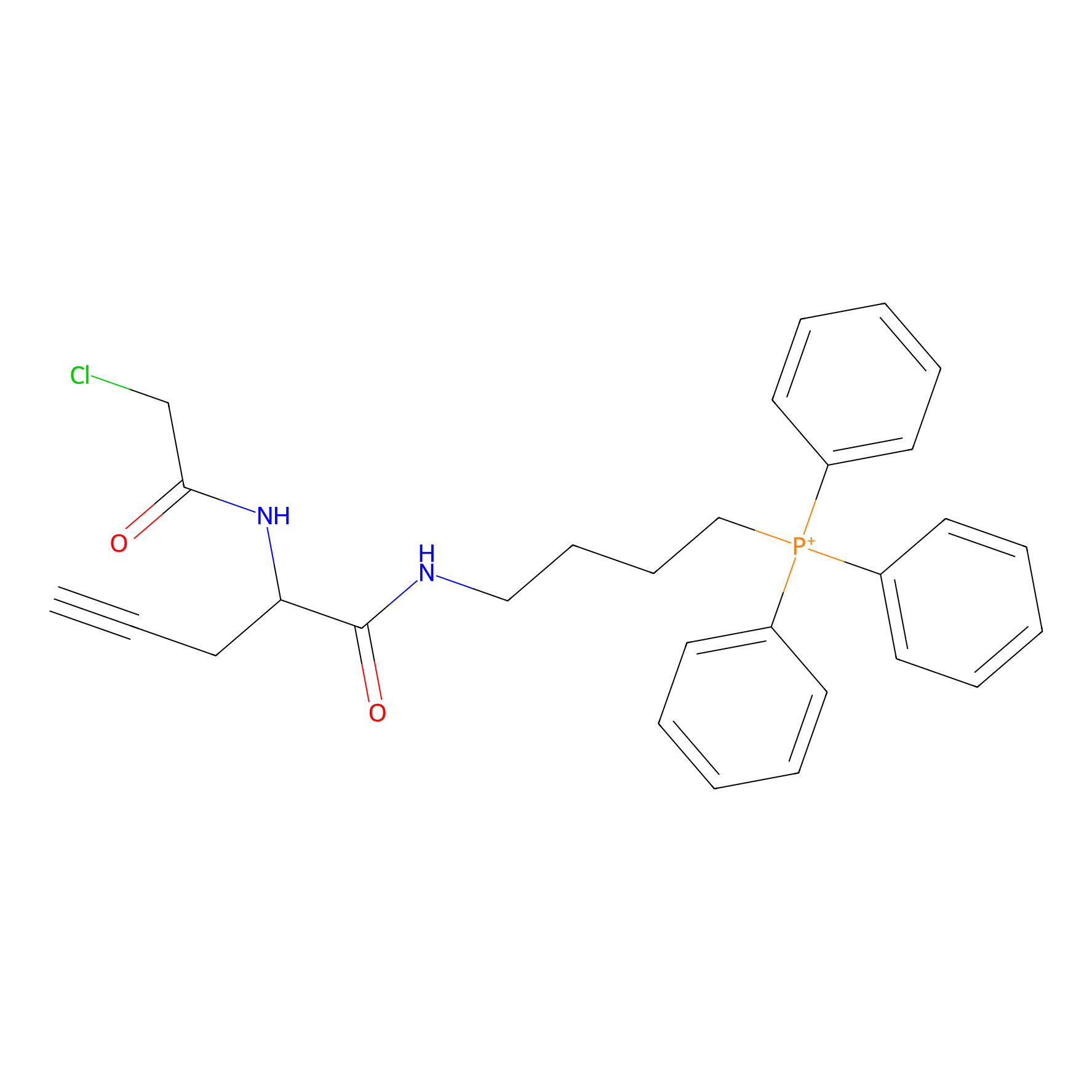 |
N.A. | LDD0427 | [18] | |
|
HHS-475 Probe Info |
 |
Y144(0.86); Y74(1.09) | LDD2238 | [10] | |
|
HHS-482 Probe Info |
 |
Y144(1.29) | LDD2239 | [10] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe12 Probe Info |
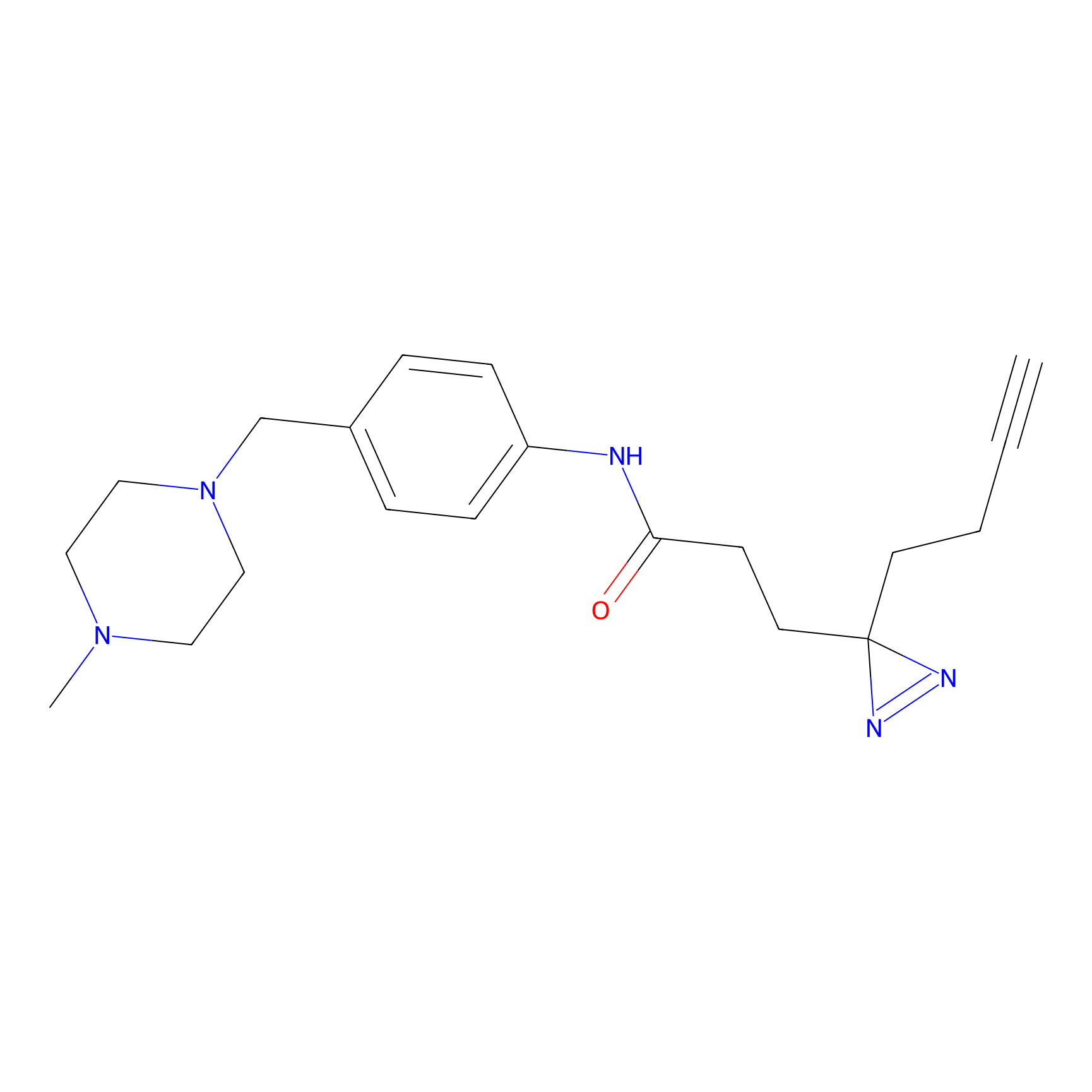 |
5.10 | LDD0473 | [19] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [20] | |
Competitor(s) Related to This Target
The Interaction Atlas With This Target
References
