Details of the Target
General Information of Target
| Target ID | LDTP01430 | |||||
|---|---|---|---|---|---|---|
| Target Name | E3 ubiquitin-protein ligase listerin (LTN1) | |||||
| Gene Name | LTN1 | |||||
| Gene ID | 26046 | |||||
| Synonyms |
C21orf10; C21orf98; KIAA0714; RNF160; ZNF294; E3 ubiquitin-protein ligase listerin; EC 2.3.2.27; RING finger protein 160; RING-type E3 ubiquitin transferase listerin; Zinc finger protein 294 |
|||||
| 3D Structure | ||||||
| Sequence |
MGGKNKQRTKGNLRPSNSGRAAELLAKEQGTVPGFIGFGTSQSDLGYVPAIQGAEEIDSL
VDSDFRMVLRKLSKKDVTTKLKAMQEFGTMCTERDTETVKGVLPYWPRIFCKISLDHDRR VREATQQAFEKLILKVKKQLAPYLKSLMGYWLMAQCDTYTPAAFAAKDAFEAAFPPSKQP EAIAFCKDEITSVLQDHLIKETPDTLSDPQTVPEEEREAKFYRVVTCSLLALKRLLCLLP DNELDSLEEKFKSLLSQNKFWKYGKHSVPQIRSAYFELVSALCQRIPQLMKEEASKVSPS VLLSIDDSDPIVCPALWEAVLYTLTTIEDCWLHVNAKKSVFPKLSTVIREGGRGLATVIY PYLLPFISKLPQSITNPKLDFFKNFLTSLVAGLSTERTKTSSLESSAVISAFFECLRFIM QQNLGEEEIEQMLVNDQLIPFIDAVLKDPGLQHGQLFNHLAETLSSWEAKADTEKDEKTA HNLENVLIHFWERLSEICVAKISEPEADVESVLGVSNLLQVLQKPKSSLKSSKKKNGKVR FADEILESNKENEKCVSSEGEKIEGWELTTEPSLTHNSSGLLSPLRKKPLEDLVCKLADI SINYVNERKSEQHLRFLSTLLDSFSSSRVFKMLLGDEKQSIVQAKPLEIAKLVQKNPAVQ FLYQKLIGWLNEDQRKDFGFLVDILYSALRCCDNDMERKKVLDDLTKVDLKWNSLLKIIE KACPSSDKHALVTPWLKGDILGEKLVNLADCLCNEDLESRVSSESHFSERWTLLSLVLSQ HVKNDYLIGDVYVERIIVRLHETLFKTKKLSEAESSDSSVSFICDVAYNYFSSAKGCLLM PSSEDLLLTLFQLCAQSKEKTHLPDFLICKLKNTWLSGVNLLVHQTDSSYKESTFLHLSA LWLKNQVQASSLDINSLQVLLSAVDDLLNTLLESEDSYLMGVYIGSVMPNDSEWEKMRQS LPMQWLHRPLLEGRLSLNYECFKTDFKEQDIKTLPSHLCTSALLSKMVLIALRKETVLEN NELEKIIAELLYSLQWCEELDNPPIFLIGFCEILQKMNITYDNLRVLGNTSGLLQLLFNR SREHGTLWSLIIAKLILSRSISSDEVKPHYKRKESFFPLTEGNLHTIQSLCPFLSKEEKK EFSAQCIPALLGWTKKDLCSTNGGFGHLAIFNSCLQTKSIDDGELLHGILKIIISWKKEH EDIFLFSCNLSEASPEVLGVNIEIIRFLSLFLKYCSSPLAESEWDFIMCSMLAWLETTSE NQALYSIPLVQLFACVSCDLACDLSAFFDSTTLDTIGNLPVNLISEWKEFFSQGIHSLLL PILVTVTGENKDVSETSFQNAMLKPMCETLTYISKEQLLSHKLPARLVADQKTNLPEYLQ TLLNTLAPLLLFRARPVQIAVYHMLYKLMPELPQYDQDNLKSYGDEEEEPALSPPAALMS LLSIQEDLLENVLGCIPVGQIVTIKPLSEDFCYVLGYLLTWKLILTFFKAASSQLRALYS MYLRKTKSLNKLLYHLFRLMPENPTYAETAVEVPNKDPKTFFTEELQLSIRETTMLPYHI PHLACSVYHMTLKDLPAMVRLWWNSSEKRVFNIVDRFTSKYVSSVLSFQEISSVQTSTQL FNGMTVKARATTREVMATYTIEDIVIELIIQLPSNYPLGSIIVESGKRVGVAVQQWRNWM LQLSTYLTHQNGSIMEGLALWKNNVDKRFEGVEDCMICFSVIHGFNYSLPKKACRTCKKK FHSACLYKWFTSSNKSTCPLCRETFF |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
LTN1 family
|
|||||
| Subcellular location |
Cytoplasm, cytosol
|
|||||
| Function |
E3 ubiquitin-protein ligase component of the ribosome quality control complex (RQC), a ribosome-associated complex that mediates ubiquitination and extraction of incompletely synthesized nascent chains for proteasomal degradation. Within the RQC complex, LTN1 is recruited to stalled 60S ribosomal subunits by NEMF and mediates ubiquitination of stalled nascent chains. Ubiquitination leads to VCP/p97 recruitment for extraction and degradation of the incomplete translation product.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| C4II | SNV: p.V486I | . | |||
| COLO792 | SNV: p.S1312F | DBIA Probe Info | |||
| HCT116 | SNV: p.V499A; p.A1744T | . | |||
| HT115 | SNV: p.R108I; p.V1217A; p.E1309Ter | DBIA Probe Info | |||
| IPC298 | SNV: p.S623F | DBIA Probe Info | |||
| K562 | Deletion: p.I199Ter | . | |||
| KARPAS422 | SNV: p.G1454R | DBIA Probe Info | |||
| KASUMI2 | Insertion: p.T479NfsTer26 | . | |||
| KMRC20 | SNV: p.T1553K | DBIA Probe Info | |||
| MCAS | SNV: p.T1640I | DBIA Probe Info | |||
| MCC13 | SNV: p.P440S | DBIA Probe Info | |||
| MDAMB453 | SNV: p.D695N | DBIA Probe Info | |||
| MFE319 | SNV: p.A1564V | DBIA Probe Info | |||
| NCIH1048 | Insertion: p.L1239FfsTer5 | . | |||
| NCIH2172 | SNV: p.E1544D | DBIA Probe Info | |||
| NCIH2286 | SNV: p.L529Ter; p.L666R; p.K1355Q | . | |||
| PC9 | Substitution: p.S531K; p.S532K | . | |||
| SKMEL30 | SNV: p.L867V | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
9.90 | LDD0402 | [1] | |
|
TH211 Probe Info |
 |
Y1514(20.00) | LDD0257 | [2] | |
|
STPyne Probe Info |
 |
K233(5.39); K609(7.18); K80(10.00) | LDD0277 | [3] | |
|
BTD Probe Info |
 |
C227(1.01) | LDD2117 | [4] | |
|
AHL-Pu-1 Probe Info |
 |
C869(2.50); C555(4.89); C237(2.91) | LDD0168 | [5] | |
|
DBIA Probe Info |
 |
C595(90.49); C981(4.91); C1146(33.70); C498(72.81) | LDD0209 | [6] | |
|
HHS-475 Probe Info |
 |
Y1514(0.62) | LDD0266 | [7] | |
|
Acrolein Probe Info |
 |
C498(0.00); C981(0.00); C237(0.00) | LDD0222 | [8] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C1131(0.00); C283(0.00); C1146(0.00); C869(0.00) | LDD0038 | [9] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [10] | |
|
Lodoacetamide azide Probe Info |
 |
C498(0.00); C999(0.00); C555(0.00); C981(0.00) | LDD0037 | [9] | |
|
IPM Probe Info |
 |
C595(0.00); C91(0.00); C498(0.00); C227(0.00) | LDD0005 | [11] | |
|
PF-06672131 Probe Info |
 |
N.A. | LDD0017 | [12] | |
|
TFBX Probe Info |
 |
C869(0.00); C999(0.00); C498(0.00); C981(0.00) | LDD0148 | [13] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [14] | |
|
AOyne Probe Info |
 |
6.60 | LDD0443 | [15] | |
|
NAIA_5 Probe Info |
 |
C91(0.00); C555(0.00); C227(0.00); C283(0.00) | LDD2223 | [16] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C280 Probe Info |
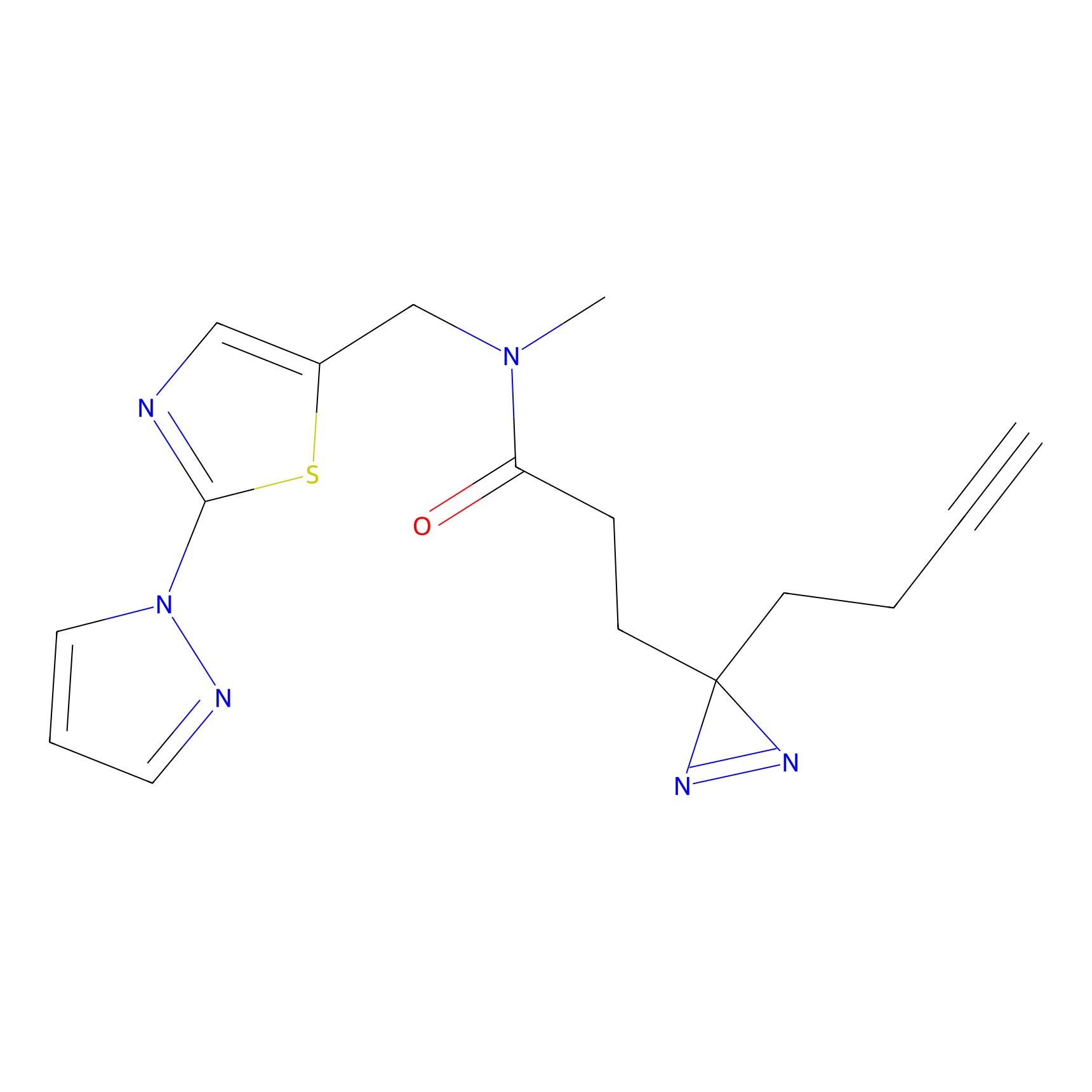 |
8.82 | LDD1950 | [17] | |
|
C293 Probe Info |
 |
21.11 | LDD1963 | [17] | |
|
C296 Probe Info |
 |
12.64 | LDD1966 | [17] | |
|
Alk-rapa Probe Info |
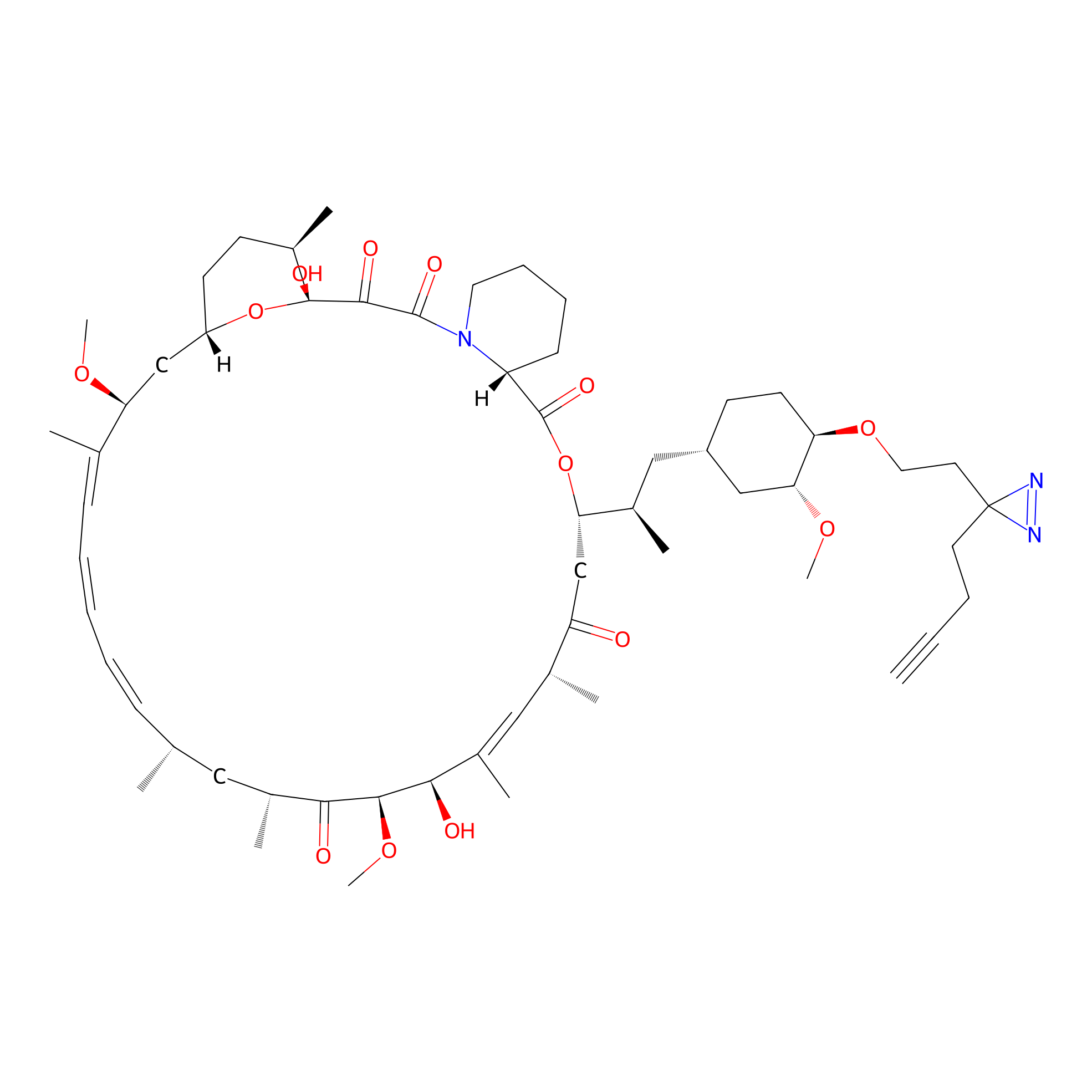 |
2.23 | LDD0213 | [18] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C227(1.01) | LDD2117 | [4] |
| LDCM0025 | 4SU-RNA | HEK-293T | C869(2.50); C555(4.89); C237(2.91) | LDD0168 | [5] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C869(2.33); C751(2.05); C555(6.56); C237(3.13) | LDD0169 | [5] |
| LDCM0214 | AC1 | HEK-293T | C723(0.99); C555(0.95); C91(1.06); C1745(0.97) | LDD1507 | [19] |
| LDCM0215 | AC10 | HEK-293T | C723(0.98); C555(1.04); C91(0.93); C1745(0.94) | LDD1508 | [19] |
| LDCM0226 | AC11 | HEK-293T | C723(0.99); C555(1.08); C91(1.01); C1745(0.94) | LDD1509 | [19] |
| LDCM0237 | AC12 | HEK-293T | C723(0.93); C555(1.00); C91(0.99); C1745(0.98) | LDD1510 | [19] |
| LDCM0259 | AC14 | HEK-293T | C723(0.99); C555(1.01); C91(0.93); C1745(0.98) | LDD1512 | [19] |
| LDCM0270 | AC15 | HEK-293T | C723(1.00); C555(0.89); C91(1.01); C498(0.94) | LDD1513 | [19] |
| LDCM0276 | AC17 | HEK-293T | C723(0.94); C555(0.97); C91(1.06); C1745(0.99) | LDD1515 | [19] |
| LDCM0277 | AC18 | HEK-293T | C723(0.98); C555(0.98); C91(1.02); C1745(1.01) | LDD1516 | [19] |
| LDCM0278 | AC19 | HEK-293T | C723(1.34); C555(1.14); C91(1.16); C1745(0.87) | LDD1517 | [19] |
| LDCM0279 | AC2 | HEK-293T | C723(0.93); C555(0.99); C91(0.97); C1745(0.96) | LDD1518 | [19] |
| LDCM0280 | AC20 | HEK-293T | C723(1.04); C555(0.98); C91(0.98); C1745(1.00) | LDD1519 | [19] |
| LDCM0281 | AC21 | HEK-293T | C723(1.05); C555(1.00); C91(0.98); C1745(0.96) | LDD1520 | [19] |
| LDCM0282 | AC22 | HEK-293T | C723(1.00); C555(1.00); C91(0.97); C1745(0.99) | LDD1521 | [19] |
| LDCM0283 | AC23 | HEK-293T | C723(1.03); C555(0.98); C91(1.00); C498(1.01) | LDD1522 | [19] |
| LDCM0284 | AC24 | HEK-293T | C723(1.00); C555(0.97); C91(1.04); C1745(0.98) | LDD1523 | [19] |
| LDCM0285 | AC25 | HEK-293T | C723(1.02); C555(0.95); C91(1.06); C1745(0.96) | LDD1524 | [19] |
| LDCM0286 | AC26 | HEK-293T | C723(0.98); C555(1.02); C91(1.06); C1745(1.00) | LDD1525 | [19] |
| LDCM0287 | AC27 | HEK-293T | C723(1.07); C555(1.01); C91(1.00); C1745(0.93) | LDD1526 | [19] |
| LDCM0288 | AC28 | HEK-293T | C723(0.98); C555(0.90); C91(0.92); C1745(1.01) | LDD1527 | [19] |
| LDCM0289 | AC29 | HEK-293T | C723(1.02); C555(1.03); C91(0.91); C1745(1.01) | LDD1528 | [19] |
| LDCM0290 | AC3 | HEK-293T | C723(0.95); C555(0.88); C91(0.92); C1745(1.00) | LDD1529 | [19] |
| LDCM0291 | AC30 | HEK-293T | C723(1.06); C555(1.07); C91(0.90); C1745(0.99) | LDD1530 | [19] |
| LDCM0292 | AC31 | HEK-293T | C723(0.98); C555(1.06); C91(1.00); C498(0.96) | LDD1531 | [19] |
| LDCM0293 | AC32 | HEK-293T | C723(1.01); C555(0.97); C91(0.95); C1745(0.97) | LDD1532 | [19] |
| LDCM0294 | AC33 | HEK-293T | C723(0.94); C555(0.95); C91(1.07); C1745(0.98) | LDD1533 | [19] |
| LDCM0295 | AC34 | HEK-293T | C723(0.94); C555(0.92); C91(1.04); C1745(0.93) | LDD1534 | [19] |
| LDCM0296 | AC35 | HEK-293T | C723(0.92); C555(0.90); C91(1.08); C1745(1.00) | LDD1535 | [19] |
| LDCM0297 | AC36 | HEK-293T | C723(0.89); C555(0.94); C91(0.96); C1745(0.98) | LDD1536 | [19] |
| LDCM0298 | AC37 | HEK-293T | C723(0.90); C555(1.01); C91(0.98); C1745(0.95) | LDD1537 | [19] |
| LDCM0299 | AC38 | HEK-293T | C723(0.94); C555(1.00); C91(0.92); C1745(0.95) | LDD1538 | [19] |
| LDCM0300 | AC39 | HEK-293T | C723(0.99); C555(0.81); C91(1.08); C498(0.96) | LDD1539 | [19] |
| LDCM0301 | AC4 | HEK-293T | C723(0.90); C555(0.97); C91(0.99); C1745(0.99) | LDD1540 | [19] |
| LDCM0302 | AC40 | HEK-293T | C723(0.83); C555(0.89); C91(0.93); C1745(1.04) | LDD1541 | [19] |
| LDCM0303 | AC41 | HEK-293T | C723(0.91); C555(0.92); C91(1.12); C1745(0.95) | LDD1542 | [19] |
| LDCM0304 | AC42 | HEK-293T | C723(0.93); C555(1.00); C91(1.07); C1745(0.93) | LDD1543 | [19] |
| LDCM0305 | AC43 | HEK-293T | C723(0.92); C555(0.94); C91(1.02); C1745(0.92) | LDD1544 | [19] |
| LDCM0306 | AC44 | HEK-293T | C723(0.89); C555(0.93); C91(0.92); C1745(0.95) | LDD1545 | [19] |
| LDCM0307 | AC45 | HEK-293T | C723(0.88); C555(1.03); C91(1.02); C1745(0.90) | LDD1546 | [19] |
| LDCM0308 | AC46 | HEK-293T | C723(0.96); C555(1.03); C91(0.94); C1745(0.99) | LDD1547 | [19] |
| LDCM0309 | AC47 | HEK-293T | C723(0.93); C555(0.77); C91(0.92); C498(0.94) | LDD1548 | [19] |
| LDCM0310 | AC48 | HEK-293T | C723(0.91); C555(0.96); C91(0.91); C1745(1.06) | LDD1549 | [19] |
| LDCM0311 | AC49 | HEK-293T | C723(0.98); C555(0.87); C91(1.16); C1745(0.95) | LDD1550 | [19] |
| LDCM0312 | AC5 | HEK-293T | C723(0.90); C555(0.98); C91(1.08); C1745(1.05) | LDD1551 | [19] |
| LDCM0313 | AC50 | HEK-293T | C723(0.92); C555(0.93); C91(0.90); C1745(0.92) | LDD1552 | [19] |
| LDCM0314 | AC51 | HEK-293T | C723(0.93); C555(0.93); C91(1.00); C1745(0.96) | LDD1553 | [19] |
| LDCM0315 | AC52 | HEK-293T | C723(0.85); C555(0.90); C91(0.92); C1745(0.97) | LDD1554 | [19] |
| LDCM0316 | AC53 | HEK-293T | C723(0.92); C555(1.01); C91(1.05); C1745(0.96) | LDD1555 | [19] |
| LDCM0317 | AC54 | HEK-293T | C723(0.87); C555(0.94); C91(0.91); C1745(0.97) | LDD1556 | [19] |
| LDCM0318 | AC55 | HEK-293T | C723(0.94); C555(0.97); C91(0.95); C498(0.91) | LDD1557 | [19] |
| LDCM0319 | AC56 | HEK-293T | C723(0.89); C555(0.94); C91(1.05); C1745(1.03) | LDD1558 | [19] |
| LDCM0320 | AC57 | HEK-293T | C723(0.99); C555(0.91); C91(1.07); C1745(0.95) | LDD1559 | [19] |
| LDCM0321 | AC58 | HEK-293T | C723(0.95); C555(0.93); C91(0.95); C1745(0.93) | LDD1560 | [19] |
| LDCM0322 | AC59 | HEK-293T | C723(0.98); C555(0.96); C91(0.89); C1745(0.97) | LDD1561 | [19] |
| LDCM0323 | AC6 | HEK-293T | C723(1.01); C555(1.08); C91(0.98); C1745(0.98) | LDD1562 | [19] |
| LDCM0324 | AC60 | HEK-293T | C723(0.98); C555(0.94); C91(1.07); C1745(0.96) | LDD1563 | [19] |
| LDCM0325 | AC61 | HEK-293T | C723(0.93); C555(1.03); C91(0.98); C1745(0.94) | LDD1564 | [19] |
| LDCM0326 | AC62 | HEK-293T | C723(0.95); C555(0.96); C91(0.91); C1745(0.99) | LDD1565 | [19] |
| LDCM0327 | AC63 | HEK-293T | C723(0.97); C555(0.93); C91(1.08); C498(0.97) | LDD1566 | [19] |
| LDCM0328 | AC64 | HEK-293T | C723(0.97); C555(0.89); C91(0.93); C1745(1.08) | LDD1567 | [19] |
| LDCM0334 | AC7 | HEK-293T | C723(0.93); C555(0.96); C91(0.94); C498(1.03) | LDD1568 | [19] |
| LDCM0345 | AC8 | HEK-293T | C723(1.01); C555(0.88); C91(1.01); C1745(1.03) | LDD1569 | [19] |
| LDCM0248 | AKOS034007472 | HEK-293T | C723(0.98); C555(1.01); C91(0.96); C1745(0.95) | LDD1511 | [19] |
| LDCM0356 | AKOS034007680 | HEK-293T | C723(0.98); C555(0.88); C91(1.01); C1745(1.00) | LDD1570 | [19] |
| LDCM0275 | AKOS034007705 | HEK-293T | C723(0.99); C555(0.97); C91(1.04); C1745(1.00) | LDD1514 | [19] |
| LDCM0108 | Chloroacetamide | HeLa | C498(0.00); C981(0.00); C237(0.00) | LDD0222 | [8] |
| LDCM0632 | CL-Sc | Hep-G2 | C595(20.00); C869(1.36); C555(0.93); C981(0.85) | LDD2227 | [16] |
| LDCM0367 | CL1 | HEK-293T | C723(0.91); C555(0.71); C91(0.90); C1745(1.12) | LDD1571 | [19] |
| LDCM0368 | CL10 | HEK-293T | C723(1.29); C555(0.79); C91(1.27); C1745(1.54) | LDD1572 | [19] |
| LDCM0369 | CL100 | HEK-293T | C723(0.95); C555(0.84); C91(1.11); C1745(1.01) | LDD1573 | [19] |
| LDCM0370 | CL101 | HEK-293T | C723(0.92); C555(0.87); C91(0.93); C1745(1.00) | LDD1574 | [19] |
| LDCM0371 | CL102 | HEK-293T | C723(1.04); C555(0.97); C91(1.04); C1745(1.03) | LDD1575 | [19] |
| LDCM0372 | CL103 | HEK-293T | C723(0.95); C555(0.94); C91(0.94); C1745(0.95) | LDD1576 | [19] |
| LDCM0373 | CL104 | HEK-293T | C723(0.96); C555(0.90); C91(1.05); C1745(1.04) | LDD1577 | [19] |
| LDCM0374 | CL105 | HEK-293T | C723(0.94); C555(1.00); C91(0.94); C1745(1.01) | LDD1578 | [19] |
| LDCM0375 | CL106 | HEK-293T | C723(1.01); C555(1.00); C91(1.04); C1745(1.05) | LDD1579 | [19] |
| LDCM0376 | CL107 | HEK-293T | C723(0.96); C555(0.99); C91(0.84); C1745(1.01) | LDD1580 | [19] |
| LDCM0377 | CL108 | HEK-293T | C723(0.91); C555(0.92); C91(0.92); C1745(1.01) | LDD1581 | [19] |
| LDCM0378 | CL109 | HEK-293T | C723(1.04); C555(0.95); C91(0.97); C1745(1.02) | LDD1582 | [19] |
| LDCM0379 | CL11 | HEK-293T | C723(0.89); C555(0.90); C91(1.13); C498(1.10) | LDD1583 | [19] |
| LDCM0380 | CL110 | HEK-293T | C723(1.12); C555(1.02); C91(1.11); C1745(1.07) | LDD1584 | [19] |
| LDCM0381 | CL111 | HEK-293T | C723(1.06); C555(0.97); C91(1.03); C1745(1.05) | LDD1585 | [19] |
| LDCM0382 | CL112 | HEK-293T | C723(0.93); C555(1.00); C91(1.07); C1745(0.99) | LDD1586 | [19] |
| LDCM0383 | CL113 | HEK-293T | C723(0.91); C555(0.86); C91(0.92); C1745(0.94) | LDD1587 | [19] |
| LDCM0384 | CL114 | HEK-293T | C723(1.13); C555(0.94); C91(1.15); C1745(1.16) | LDD1588 | [19] |
| LDCM0385 | CL115 | HEK-293T | C723(0.89); C555(0.80); C91(0.84); C1745(1.01) | LDD1589 | [19] |
| LDCM0386 | CL116 | HEK-293T | C723(0.95); C555(0.90); C91(0.94); C1745(0.99) | LDD1590 | [19] |
| LDCM0387 | CL117 | HEK-293T | C723(0.89); C555(0.96); C91(0.97); C1745(0.98) | LDD1591 | [19] |
| LDCM0388 | CL118 | HEK-293T | C723(0.98); C555(0.99); C91(1.15); C1745(0.93) | LDD1592 | [19] |
| LDCM0389 | CL119 | HEK-293T | C723(0.86); C555(0.89); C91(0.79); C1745(0.93) | LDD1593 | [19] |
| LDCM0390 | CL12 | HEK-293T | C723(1.03); C555(0.85); C91(1.03); C1745(0.82) | LDD1594 | [19] |
| LDCM0391 | CL120 | HEK-293T | C723(0.99); C555(0.92); C91(0.98); C1745(0.98) | LDD1595 | [19] |
| LDCM0392 | CL121 | HEK-293T | C723(0.99); C555(0.90); C91(0.92); C1745(0.97) | LDD1596 | [19] |
| LDCM0393 | CL122 | HEK-293T | C723(1.00); C555(1.00); C91(1.02); C1745(1.07) | LDD1597 | [19] |
| LDCM0394 | CL123 | HEK-293T | C723(1.64); C555(0.84); C91(0.89); C1745(1.20) | LDD1598 | [19] |
| LDCM0395 | CL124 | HEK-293T | C723(0.98); C555(0.95); C91(0.99); C1745(1.00) | LDD1599 | [19] |
| LDCM0396 | CL125 | HEK-293T | C723(0.95); C555(0.90); C91(0.96); C1745(0.93) | LDD1600 | [19] |
| LDCM0397 | CL126 | HEK-293T | C723(1.01); C555(1.00); C91(0.97); C1745(0.97) | LDD1601 | [19] |
| LDCM0398 | CL127 | HEK-293T | C723(0.97); C555(0.91); C91(1.03); C1745(0.97) | LDD1602 | [19] |
| LDCM0399 | CL128 | HEK-293T | C723(0.93); C555(0.98); C91(0.89); C1745(0.98) | LDD1603 | [19] |
| LDCM0400 | CL13 | HEK-293T | C723(0.89); C555(0.86); C91(0.97); C1745(0.92) | LDD1604 | [19] |
| LDCM0401 | CL14 | HEK-293T | C723(0.92); C555(0.91); C91(1.00); C1745(0.94) | LDD1605 | [19] |
| LDCM0402 | CL15 | HEK-293T | C723(1.43); C555(0.78); C91(1.14); C1745(1.10) | LDD1606 | [19] |
| LDCM0403 | CL16 | HEK-293T | C723(0.94); C555(0.90); C91(0.95); C1745(0.97) | LDD1607 | [19] |
| LDCM0404 | CL17 | HEK-293T | C723(1.49); C555(0.77); C91(1.14); C1745(1.17) | LDD1608 | [19] |
| LDCM0405 | CL18 | HEK-293T | C723(0.91); C555(0.93); C91(1.02); C1745(0.94) | LDD1609 | [19] |
| LDCM0406 | CL19 | HEK-293T | C723(0.88); C555(0.95); C91(1.08); C1745(1.00) | LDD1610 | [19] |
| LDCM0407 | CL2 | HEK-293T | C723(0.94); C555(0.92); C91(1.11); C1745(1.01) | LDD1611 | [19] |
| LDCM0408 | CL20 | HEK-293T | C723(0.98); C555(1.11); C91(1.12); C1745(1.02) | LDD1612 | [19] |
| LDCM0409 | CL21 | HEK-293T | C723(1.14); C555(1.06); C91(1.16); C1745(1.24) | LDD1613 | [19] |
| LDCM0410 | CL22 | HEK-293T | C723(0.82); C555(0.89); C91(1.07); C1745(1.11) | LDD1614 | [19] |
| LDCM0411 | CL23 | HEK-293T | C723(0.81); C555(0.91); C91(1.13); C498(1.07) | LDD1615 | [19] |
| LDCM0412 | CL24 | HEK-293T | C723(0.94); C555(0.91); C91(0.96); C1745(0.79) | LDD1616 | [19] |
| LDCM0413 | CL25 | HEK-293T | C723(1.21); C555(0.91); C91(0.90); C1745(1.20) | LDD1617 | [19] |
| LDCM0414 | CL26 | HEK-293T | C723(0.96); C555(1.01); C91(1.05); C1745(1.07) | LDD1618 | [19] |
| LDCM0415 | CL27 | HEK-293T | C723(0.95); C555(0.95); C91(0.92); C1745(1.02) | LDD1619 | [19] |
| LDCM0416 | CL28 | HEK-293T | C723(1.07); C555(0.90); C91(0.96); C1745(1.02) | LDD1620 | [19] |
| LDCM0417 | CL29 | HEK-293T | C723(0.97); C555(0.93); C91(1.13); C1745(1.02) | LDD1621 | [19] |
| LDCM0418 | CL3 | HEK-293T | C723(0.97); C555(0.79); C91(0.94); C1745(1.02) | LDD1622 | [19] |
| LDCM0419 | CL30 | HEK-293T | C723(0.91); C555(0.97); C91(1.01); C1745(0.99) | LDD1623 | [19] |
| LDCM0420 | CL31 | HEK-293T | C723(1.00); C555(1.09); C91(0.97); C1745(1.12) | LDD1624 | [19] |
| LDCM0421 | CL32 | HEK-293T | C723(0.93); C555(1.00); C91(0.96); C1745(1.08) | LDD1625 | [19] |
| LDCM0422 | CL33 | HEK-293T | C723(1.90); C555(0.93); C91(1.19); C1745(1.38) | LDD1626 | [19] |
| LDCM0423 | CL34 | HEK-293T | C723(1.00); C555(1.03); C91(1.13); C1745(1.32) | LDD1627 | [19] |
| LDCM0424 | CL35 | HEK-293T | C723(0.90); C555(1.13); C91(1.09); C498(1.05) | LDD1628 | [19] |
| LDCM0425 | CL36 | HEK-293T | C723(0.96); C555(1.09); C91(1.00); C1745(0.72) | LDD1629 | [19] |
| LDCM0426 | CL37 | HEK-293T | C723(0.93); C555(1.05); C91(0.90); C1745(1.03) | LDD1630 | [19] |
| LDCM0428 | CL39 | HEK-293T | C723(0.96); C555(0.89); C91(0.96); C1745(1.16) | LDD1632 | [19] |
| LDCM0429 | CL4 | HEK-293T | C723(1.06); C555(0.82); C91(1.08); C1745(1.10) | LDD1633 | [19] |
| LDCM0430 | CL40 | HEK-293T | C723(0.97); C555(0.97); C91(1.23); C1745(0.97) | LDD1634 | [19] |
| LDCM0431 | CL41 | HEK-293T | C723(0.92); C555(0.85); C91(0.95); C1745(1.07) | LDD1635 | [19] |
| LDCM0432 | CL42 | HEK-293T | C723(0.92); C555(0.99); C91(0.93); C1745(0.96) | LDD1636 | [19] |
| LDCM0433 | CL43 | HEK-293T | C723(1.01); C555(0.96); C91(1.03); C1745(1.05) | LDD1637 | [19] |
| LDCM0434 | CL44 | HEK-293T | C723(0.91); C555(1.02); C91(1.05); C1745(1.03) | LDD1638 | [19] |
| LDCM0435 | CL45 | HEK-293T | C723(1.06); C555(1.01); C91(1.14); C1745(1.16) | LDD1639 | [19] |
| LDCM0436 | CL46 | HEK-293T | C723(0.97); C555(1.02); C91(1.09); C1745(1.17) | LDD1640 | [19] |
| LDCM0437 | CL47 | HEK-293T | C723(0.89); C555(0.94); C91(1.12); C498(1.05) | LDD1641 | [19] |
| LDCM0438 | CL48 | HEK-293T | C723(0.91); C555(0.98); C91(1.11); C1745(0.73) | LDD1642 | [19] |
| LDCM0439 | CL49 | HEK-293T | C723(0.91); C555(0.76); C91(0.92); C1745(0.99) | LDD1643 | [19] |
| LDCM0440 | CL5 | HEK-293T | C723(0.92); C555(0.84); C91(1.04); C1745(1.03) | LDD1644 | [19] |
| LDCM0441 | CL50 | HEK-293T | C723(1.02); C555(0.96); C91(1.03); C1745(1.06) | LDD1645 | [19] |
| LDCM0443 | CL52 | HEK-293T | C723(0.87); C555(0.90); C91(0.97); C1745(1.02) | LDD1646 | [19] |
| LDCM0444 | CL53 | HEK-293T | C723(1.03); C555(0.80); C91(1.24); C1745(1.14) | LDD1647 | [19] |
| LDCM0445 | CL54 | HEK-293T | C723(1.64); C555(0.85); C91(0.98); C1745(1.27) | LDD1648 | [19] |
| LDCM0446 | CL55 | HEK-293T | C723(0.93); C555(0.97); C91(1.05); C1745(1.01) | LDD1649 | [19] |
| LDCM0447 | CL56 | HEK-293T | C723(0.85); C555(0.90); C91(0.99); C1745(1.09) | LDD1650 | [19] |
| LDCM0448 | CL57 | HEK-293T | C723(1.18); C555(1.04); C91(1.09); C1745(1.25) | LDD1651 | [19] |
| LDCM0449 | CL58 | HEK-293T | C723(0.87); C555(0.98); C91(1.15); C1745(1.31) | LDD1652 | [19] |
| LDCM0450 | CL59 | HEK-293T | C723(0.83); C555(0.89); C91(1.03); C498(1.05) | LDD1653 | [19] |
| LDCM0451 | CL6 | HEK-293T | C723(1.04); C555(0.83); C91(1.09); C1745(1.08) | LDD1654 | [19] |
| LDCM0452 | CL60 | HEK-293T | C723(0.97); C555(0.91); C91(0.97); C1745(0.79) | LDD1655 | [19] |
| LDCM0453 | CL61 | HEK-293T | C723(0.88); C555(0.76); C91(0.90); C1745(0.90) | LDD1656 | [19] |
| LDCM0454 | CL62 | HEK-293T | C723(0.93); C555(1.00); C91(0.92); C1745(0.94) | LDD1657 | [19] |
| LDCM0455 | CL63 | HEK-293T | C723(0.92); C555(0.91); C91(0.83); C1745(0.93) | LDD1658 | [19] |
| LDCM0456 | CL64 | HEK-293T | C723(1.08); C555(0.86); C91(1.12); C1745(1.13) | LDD1659 | [19] |
| LDCM0457 | CL65 | HEK-293T | C723(1.02); C555(0.92); C91(0.98); C1745(0.95) | LDD1660 | [19] |
| LDCM0458 | CL66 | HEK-293T | C723(1.05); C555(0.97); C91(1.10); C1745(0.94) | LDD1661 | [19] |
| LDCM0459 | CL67 | HEK-293T | C723(0.97); C555(0.96); C91(1.01); C1745(0.98) | LDD1662 | [19] |
| LDCM0460 | CL68 | HEK-293T | C723(0.98); C555(0.95); C91(1.02); C1745(1.08) | LDD1663 | [19] |
| LDCM0461 | CL69 | HEK-293T | C723(1.06); C555(1.11); C91(1.00); C1745(1.03) | LDD1664 | [19] |
| LDCM0462 | CL7 | HEK-293T | C723(0.88); C555(0.88); C91(1.07); C1745(1.01) | LDD1665 | [19] |
| LDCM0463 | CL70 | HEK-293T | C723(0.84); C555(0.98); C91(1.15); C1745(1.10) | LDD1666 | [19] |
| LDCM0464 | CL71 | HEK-293T | C723(0.79); C555(1.07); C91(0.96); C498(0.97) | LDD1667 | [19] |
| LDCM0465 | CL72 | HEK-293T | C723(0.99); C555(0.99); C91(1.12); C1745(0.82) | LDD1668 | [19] |
| LDCM0466 | CL73 | HEK-293T | C723(0.94); C555(0.79); C91(1.01); C1745(1.09) | LDD1669 | [19] |
| LDCM0467 | CL74 | HEK-293T | C723(0.94); C555(0.95); C91(1.00); C1745(0.96) | LDD1670 | [19] |
| LDCM0469 | CL76 | HEK-293T | C723(1.00); C555(0.89); C91(0.95); C1745(0.98) | LDD1672 | [19] |
| LDCM0470 | CL77 | HEK-293T | C723(1.07); C555(0.85); C91(1.12); C1745(1.09) | LDD1673 | [19] |
| LDCM0471 | CL78 | HEK-293T | C723(0.96); C555(0.95); C91(0.94); C1745(0.96) | LDD1674 | [19] |
| LDCM0472 | CL79 | HEK-293T | C723(0.94); C555(1.02); C91(0.95); C1745(1.08) | LDD1675 | [19] |
| LDCM0473 | CL8 | HEK-293T | C723(3.07); C555(0.78); C91(1.68); C1745(1.35) | LDD1676 | [19] |
| LDCM0474 | CL80 | HEK-293T | C723(0.86); C555(0.87); C91(1.05); C1745(1.02) | LDD1677 | [19] |
| LDCM0475 | CL81 | HEK-293T | C723(0.90); C555(1.14); C91(0.91); C1745(1.04) | LDD1678 | [19] |
| LDCM0476 | CL82 | HEK-293T | C723(0.80); C555(0.87); C91(1.01); C1745(1.08) | LDD1679 | [19] |
| LDCM0477 | CL83 | HEK-293T | C723(0.86); C555(0.93); C91(1.09); C498(0.98) | LDD1680 | [19] |
| LDCM0478 | CL84 | HEK-293T | C723(0.99); C555(0.95); C91(1.11); C1745(0.92) | LDD1681 | [19] |
| LDCM0479 | CL85 | HEK-293T | C723(0.88); C555(0.82); C91(0.93); C1745(0.95) | LDD1682 | [19] |
| LDCM0480 | CL86 | HEK-293T | C723(0.92); C555(0.98); C91(0.99); C1745(0.95) | LDD1683 | [19] |
| LDCM0481 | CL87 | HEK-293T | C723(0.91); C555(0.92); C91(0.81); C1745(0.93) | LDD1684 | [19] |
| LDCM0482 | CL88 | HEK-293T | C723(0.95); C555(0.87); C91(1.13); C1745(0.98) | LDD1685 | [19] |
| LDCM0483 | CL89 | HEK-293T | C723(0.96); C555(0.85); C91(1.21); C1745(0.94) | LDD1686 | [19] |
| LDCM0484 | CL9 | HEK-293T | C723(0.94); C555(1.04); C91(1.09); C1745(1.17) | LDD1687 | [19] |
| LDCM0485 | CL90 | HEK-293T | C723(2.44); C555(0.81); C91(1.23); C1745(1.35) | LDD1688 | [19] |
| LDCM0486 | CL91 | HEK-293T | C723(0.94); C555(0.85); C91(0.92); C1745(1.03) | LDD1689 | [19] |
| LDCM0487 | CL92 | HEK-293T | C723(1.02); C555(0.92); C91(0.97); C1745(1.11) | LDD1690 | [19] |
| LDCM0488 | CL93 | HEK-293T | C723(1.20); C555(0.96); C91(1.08); C1745(1.15) | LDD1691 | [19] |
| LDCM0489 | CL94 | HEK-293T | C723(0.91); C555(0.98); C91(0.92); C1745(1.11) | LDD1692 | [19] |
| LDCM0490 | CL95 | HEK-293T | C723(1.34); C555(0.81); C91(1.34); C498(1.07) | LDD1693 | [19] |
| LDCM0491 | CL96 | HEK-293T | C723(0.96); C555(0.88); C91(1.07); C1745(1.03) | LDD1694 | [19] |
| LDCM0492 | CL97 | HEK-293T | C723(0.93); C555(0.80); C91(1.06); C1745(1.09) | LDD1695 | [19] |
| LDCM0493 | CL98 | HEK-293T | C723(0.97); C555(0.96); C91(1.00); C1745(1.05) | LDD1696 | [19] |
| LDCM0494 | CL99 | HEK-293T | C723(0.93); C555(0.90); C91(0.90); C1745(1.00) | LDD1697 | [19] |
| LDCM0634 | CY-0357 | Hep-G2 | C869(0.31) | LDD2228 | [16] |
| LDCM0495 | E2913 | HEK-293T | C723(0.83); C555(0.82); C91(0.91); C1745(0.93) | LDD1698 | [19] |
| LDCM0204 | EV-97 | T cell | C723(20.00) | LDD0525 | [20] |
| LDCM0625 | F8 | Ramos | C869(1.20) | LDD2187 | [21] |
| LDCM0573 | Fragment11 | Ramos | C498(8.97); C869(20.00) | LDD2190 | [21] |
| LDCM0586 | Fragment28 | Ramos | C869(1.23) | LDD2198 | [21] |
| LDCM0468 | Fragment33 | HEK-293T | C723(0.87); C555(0.91); C91(0.82); C1745(1.01) | LDD1671 | [19] |
| LDCM0427 | Fragment51 | HEK-293T | C723(0.93); C555(1.03); C91(1.14); C1745(0.92) | LDD1631 | [19] |
| LDCM0569 | Fragment7 | Ramos | C498(0.81); C869(3.12) | LDD2186 | [21] |
| LDCM0118 | HHS-0301 | DM93 | Y1514(0.62) | LDD0266 | [7] |
| LDCM0120 | HHS-0701 | DM93 | Y1514(0.31) | LDD0268 | [7] |
| LDCM0022 | KB02 | HEK-293T | C723(0.88); C555(0.90); C91(1.00); C595(0.92) | LDD1492 | [19] |
| LDCM0023 | KB03 | Jurkat | C595(90.49); C981(4.91); C1146(33.70); C498(72.81) | LDD0209 | [6] |
| LDCM0024 | KB05 | G361 | C769(9.42); C232(3.11) | LDD3311 | [22] |
| LDCM0090 | Rapamycin | JHH-7 | 2.23 | LDD0213 | [18] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Stimulator of interferon genes protein (STING1) | STING family | Q86WV6 | |||
| Toll/interleukin-1 receptor domain-containing adapter protein (TIRAP) | . | P58753 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Interferon regulatory factor 7 (IRF7) | IRF family | Q92985 | |||
References
