Details of the Target
General Information of Target
| Target ID | LDTP14068 | |||||
|---|---|---|---|---|---|---|
| Target Name | Cysteine protease ATG4B (ATG4B) | |||||
| Gene Name | ATG4B | |||||
| Gene ID | 23192 | |||||
| Synonyms |
APG4B; AUTL1; KIAA0943; Cysteine protease ATG4B; EC 3.4.22.-; AUT-like 1 cysteine endopeptidase; Autophagy-related cysteine endopeptidase 1; Autophagin-1; Autophagy-related protein 4 homolog B; HsAPG4B; hAPG4B
|
|||||
| 3D Structure | ||||||
| Sequence |
MAEASAAGADSGAAVAAHRFFCHFCKGEVSPKLPEYICPRCESGFIEEVTDDSSFLGGGG
SRIDNTTTTHFAELWGHLDHTMFFQDFRPFLSSSPLDQDNRANERGHQTHTDFWGARPPR LPLGRRYRSRGSSRPDRSPAIEGILQHIFAGFFANSAIPGSPHPFSWSGMLHSNPGDYAW GQTGLDAIVTQLLGQLENTGPPPADKEKITSLPTVTVTQEQVDMGLECPVCKEDYTVEEE VRQLPCNHFFHSSCIVPWLELHDTCPVCRKSLNGEDSTRQSQSTEASASNRFSNDSQLHD RWTF |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Peptidase C54 family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Cysteine protease that plays a key role in autophagy by mediating both proteolytic activation and delipidation of ATG8 family proteins . Required for canonical autophagy (macroautophagy), non-canonical autophagy as well as for mitophagy. The protease activity is required for proteolytic activation of ATG8 family proteins: cleaves the C-terminal amino acid of ATG8 proteins MAP1LC3A, MAP1LC3B, MAP1LC3C, GABARAPL1, GABARAPL2 and GABARAP, to reveal a C-terminal glycine. Exposure of the glycine at the C-terminus is essential for ATG8 proteins conjugation to phosphatidylethanolamine (PE) and insertion to membranes, which is necessary for autophagy . Protease activity is also required to counteract formation of high-molecular weight conjugates of ATG8 proteins (ATG8ylation): acts as a deubiquitinating-like enzyme that removes ATG8 conjugated to other proteins, such as ATG3. In addition to the protease activity, also mediates delipidation of ATG8 family proteins . Catalyzes delipidation of PE-conjugated forms of ATG8 proteins during macroautophagy. Also involved in non-canonical autophagy, a parallel pathway involving conjugation of ATG8 proteins to single membranes at endolysosomal compartments, by catalyzing delipidation of ATG8 proteins conjugated to phosphatidylserine (PS). Compared to other members of the family (ATG4A, ATG4C or ATG4C), constitutes the major protein for proteolytic activation of ATG8 proteins, while it displays weaker delipidation activity than other ATG4 paralogs. Involved in phagophore growth during mitophagy independently of its protease activity and of ATG8 proteins: acts by regulating ATG9A trafficking to mitochondria and promoting phagophore-endoplasmic reticulum contacts during the lipid transfer phase of mitophagy.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| AN3CA | SNV: p.H300R | DBIA Probe Info | |||
| HCC1187 | Insertion: p.D200IfsTer46 | DBIA Probe Info | |||
| HEP3B217 | SNV: p.A114T | . | |||
| KO52 | SNV: p.F16L | DBIA Probe Info | |||
| NCIH1793 | Deletion: p.I42TfsTer40 | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
IPM Probe Info |
 |
C333(0.00); C78(0.00); C89(0.00); C189(0.00) | LDD0241 | [2] | |
|
DBIA Probe Info |
 |
C277(1.34) | LDD3314 | [3] | |
|
DA-P3 Probe Info |
 |
11.07 | LDD0179 | [4] | |
|
AHL-Pu-1 Probe Info |
 |
C361(6.95); C189(4.86) | LDD0168 | [5] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0222 | [6] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C361(0.00); C323(0.00); C74(0.00); C189(0.00) | LDD0038 | [7] | |
|
IA-alkyne Probe Info |
 |
C203(0.00); C361(0.00); C323(0.00); C74(0.00) | LDD0036 | [7] | |
|
Lodoacetamide azide Probe Info |
 |
C361(0.00); C323(0.00); C74(0.00); C333(0.00) | LDD0037 | [7] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [8] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [9] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [9] | |
|
ENE Probe Info |
 |
N.A. | LDD0006 | [10] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [10] | |
|
W1 Probe Info |
 |
C189(0.00); C333(0.00); C74(0.00) | LDD0236 | [2] | |
|
AOyne Probe Info |
 |
14.20 | LDD0443 | [11] | |
|
NAIA_5 Probe Info |
 |
C203(0.00); C323(0.00); C333(0.00) | LDD2223 | [8] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C320 Probe Info |
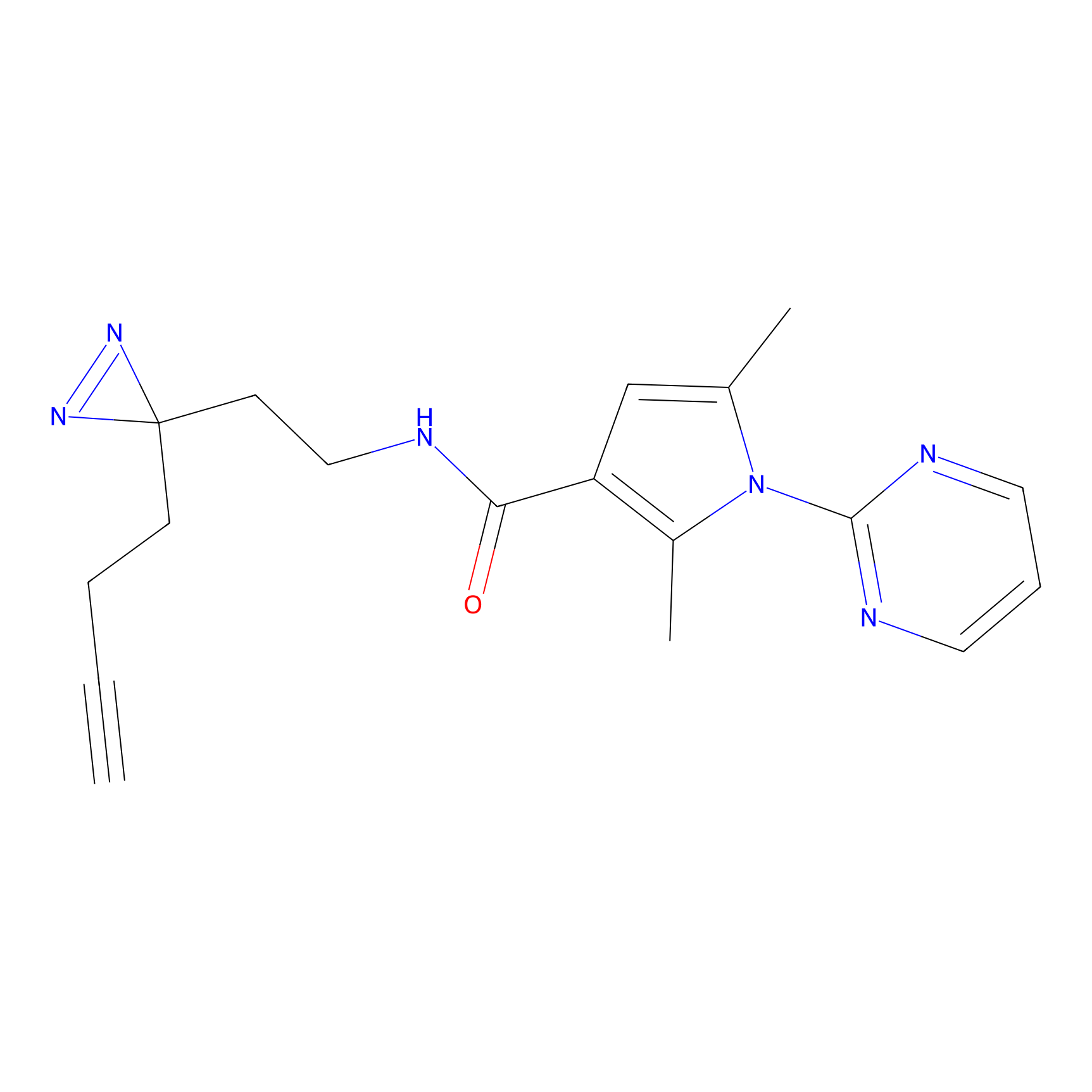 |
8.06 | LDD1986 | [12] | |
|
C322 Probe Info |
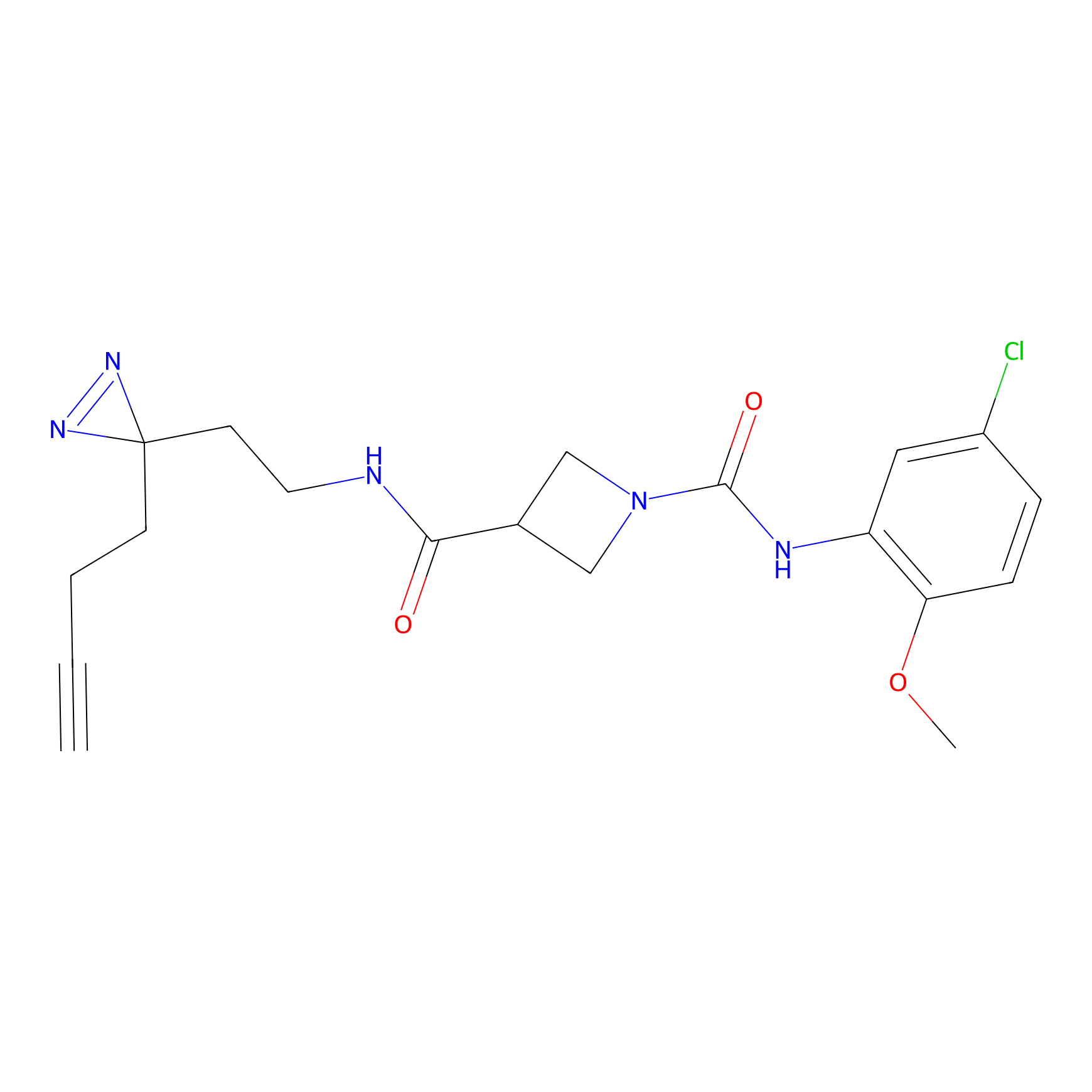 |
6.68 | LDD1988 | [12] | |
|
C326 Probe Info |
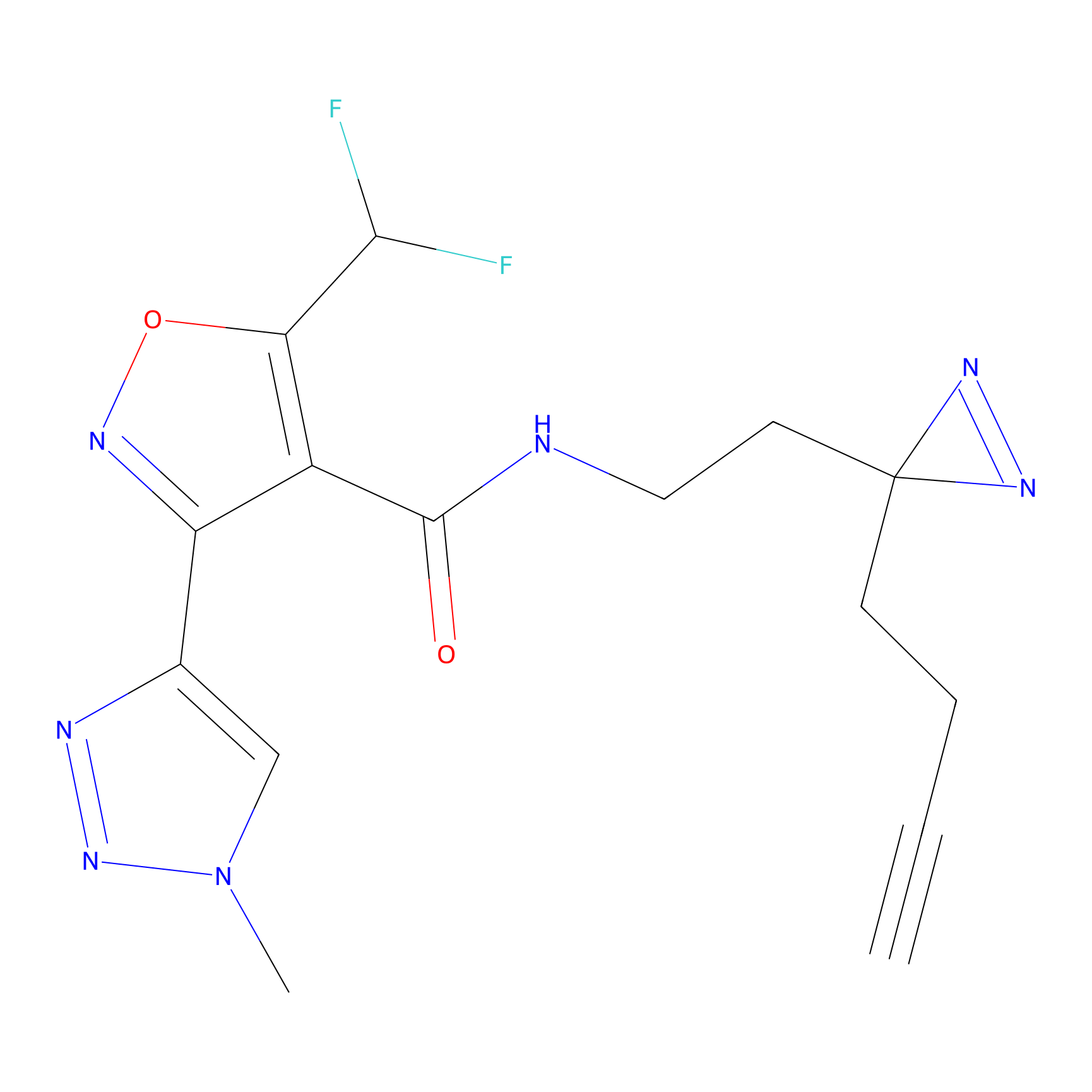 |
7.41 | LDD1990 | [12] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | HEK-293T | C361(6.95); C189(4.86) | LDD0168 | [5] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C333(2.68); C189(3.46) | LDD0169 | [5] |
| LDCM0214 | AC1 | HEK-293T | C203(1.23); C74(1.00); C323(0.93); C333(0.88) | LDD1507 | [13] |
| LDCM0215 | AC10 | HEK-293T | C203(0.97); C74(1.11); C323(1.00); C333(1.05) | LDD1508 | [13] |
| LDCM0226 | AC11 | HEK-293T | C203(0.90); C74(1.03); C323(1.09); C333(1.08) | LDD1509 | [13] |
| LDCM0237 | AC12 | HEK-293T | C203(1.05); C74(1.01); C323(1.00); C333(1.07) | LDD1510 | [13] |
| LDCM0259 | AC14 | HEK-293T | C203(1.09); C74(1.14); C323(0.98); C189(1.15) | LDD1512 | [13] |
| LDCM0270 | AC15 | HEK-293T | C203(0.98); C74(1.02); C323(0.96); C333(1.11) | LDD1513 | [13] |
| LDCM0276 | AC17 | HEK-293T | C203(1.10); C74(1.06); C323(0.93); C333(1.02) | LDD1515 | [13] |
| LDCM0277 | AC18 | HEK-293T | C203(1.07); C74(0.97); C323(0.94); C333(1.06) | LDD1516 | [13] |
| LDCM0278 | AC19 | HEK-293T | C203(1.06); C74(1.00); C323(0.98); C333(0.94) | LDD1517 | [13] |
| LDCM0279 | AC2 | HEK-293T | C203(0.99); C74(1.05); C323(1.11); C333(1.03) | LDD1518 | [13] |
| LDCM0280 | AC20 | HEK-293T | C203(1.10); C74(0.99); C323(0.99); C333(1.06) | LDD1519 | [13] |
| LDCM0281 | AC21 | HEK-293T | C203(1.09); C74(1.01); C323(1.00); C189(1.19) | LDD1520 | [13] |
| LDCM0282 | AC22 | HEK-293T | C203(0.92); C74(1.01); C323(0.97); C189(1.01) | LDD1521 | [13] |
| LDCM0283 | AC23 | HEK-293T | C203(0.95); C74(1.04); C323(0.96); C333(1.06) | LDD1522 | [13] |
| LDCM0284 | AC24 | HEK-293T | C203(1.02); C74(1.11); C323(0.99); C333(1.03) | LDD1523 | [13] |
| LDCM0285 | AC25 | HEK-293T | C203(1.17); C74(1.18); C323(0.93); C333(0.85) | LDD1524 | [13] |
| LDCM0286 | AC26 | HEK-293T | C203(0.99); C74(1.07); C323(1.03); C333(1.02) | LDD1525 | [13] |
| LDCM0287 | AC27 | HEK-293T | C203(1.01); C74(1.00); C323(1.02); C333(1.05) | LDD1526 | [13] |
| LDCM0288 | AC28 | HEK-293T | C203(0.96); C74(0.96); C323(0.91); C333(0.99) | LDD1527 | [13] |
| LDCM0289 | AC29 | HEK-293T | C203(0.92); C74(1.01); C323(0.97); C189(1.06) | LDD1528 | [13] |
| LDCM0290 | AC3 | HEK-293T | C203(0.95); C74(1.01); C323(1.04); C333(0.93) | LDD1529 | [13] |
| LDCM0291 | AC30 | HEK-293T | C203(0.87); C74(1.11); C323(0.96); C189(0.96) | LDD1530 | [13] |
| LDCM0292 | AC31 | HEK-293T | C203(0.94); C74(1.07); C323(1.04); C333(0.96) | LDD1531 | [13] |
| LDCM0293 | AC32 | HEK-293T | C203(1.09); C74(1.11); C323(1.09); C333(1.04) | LDD1532 | [13] |
| LDCM0294 | AC33 | HEK-293T | C203(1.13); C74(1.07); C323(0.96); C333(0.91) | LDD1533 | [13] |
| LDCM0295 | AC34 | HEK-293T | C203(1.13); C74(1.07); C323(1.11); C333(0.94) | LDD1534 | [13] |
| LDCM0296 | AC35 | HEK-293T | C203(0.93); C74(1.06); C323(1.03); C333(0.88) | LDD1535 | [13] |
| LDCM0297 | AC36 | HEK-293T | C203(1.02); C74(1.01); C323(1.10); C333(1.08) | LDD1536 | [13] |
| LDCM0298 | AC37 | HEK-293T | C203(0.97); C74(0.98); C323(0.98); C189(1.15) | LDD1537 | [13] |
| LDCM0299 | AC38 | HEK-293T | C203(1.02); C74(1.04); C323(0.97); C189(1.10) | LDD1538 | [13] |
| LDCM0300 | AC39 | HEK-293T | C203(1.08); C74(1.09); C323(0.94); C333(1.03) | LDD1539 | [13] |
| LDCM0301 | AC4 | HEK-293T | C203(0.99); C74(0.99); C323(1.11); C333(1.06) | LDD1540 | [13] |
| LDCM0302 | AC40 | HEK-293T | C203(1.12); C74(1.11); C323(1.03); C333(0.96) | LDD1541 | [13] |
| LDCM0303 | AC41 | HEK-293T | C203(1.30); C74(1.04); C323(0.97); C333(0.98) | LDD1542 | [13] |
| LDCM0304 | AC42 | HEK-293T | C203(1.10); C74(1.14); C323(1.03); C333(0.98) | LDD1543 | [13] |
| LDCM0305 | AC43 | HEK-293T | C203(1.02); C74(1.07); C323(1.00); C333(1.03) | LDD1544 | [13] |
| LDCM0306 | AC44 | HEK-293T | C203(1.02); C74(1.01); C323(1.05); C333(1.19) | LDD1545 | [13] |
| LDCM0307 | AC45 | HEK-293T | C203(0.86); C74(1.00); C323(0.98); C189(1.15) | LDD1546 | [13] |
| LDCM0308 | AC46 | HEK-293T | C203(0.97); C74(1.18); C323(1.03); C189(1.29) | LDD1547 | [13] |
| LDCM0309 | AC47 | HEK-293T | C203(0.89); C74(1.04); C323(1.03); C333(1.19) | LDD1548 | [13] |
| LDCM0310 | AC48 | HEK-293T | C203(0.98); C74(1.16); C323(0.97); C333(1.07) | LDD1549 | [13] |
| LDCM0311 | AC49 | HEK-293T | C203(1.22); C74(1.25); C323(0.97); C333(0.90) | LDD1550 | [13] |
| LDCM0312 | AC5 | HEK-293T | C203(0.99); C74(1.01); C323(0.96); C189(1.21) | LDD1551 | [13] |
| LDCM0313 | AC50 | HEK-293T | C203(1.03); C74(1.13); C323(1.04); C333(1.01) | LDD1552 | [13] |
| LDCM0314 | AC51 | HEK-293T | C203(0.94); C74(1.06); C323(1.10); C333(1.08) | LDD1553 | [13] |
| LDCM0315 | AC52 | HEK-293T | C203(1.06); C74(1.04); C323(1.06); C333(1.25) | LDD1554 | [13] |
| LDCM0316 | AC53 | HEK-293T | C203(0.87); C74(0.98); C323(0.97); C189(1.15) | LDD1555 | [13] |
| LDCM0317 | AC54 | HEK-293T | C203(0.96); C74(1.15); C323(1.00); C189(1.22) | LDD1556 | [13] |
| LDCM0318 | AC55 | HEK-293T | C203(0.88); C74(1.07); C323(1.00); C333(0.93) | LDD1557 | [13] |
| LDCM0319 | AC56 | HEK-293T | C203(1.06); C74(1.20); C323(1.04); C333(1.02) | LDD1558 | [13] |
| LDCM0320 | AC57 | HEK-293T | C203(1.18); C74(1.03); C323(0.98); C333(0.93) | LDD1559 | [13] |
| LDCM0321 | AC58 | HEK-293T | C203(0.95); C74(1.06); C323(1.11); C333(1.09) | LDD1560 | [13] |
| LDCM0322 | AC59 | HEK-293T | C203(1.10); C74(1.05); C323(1.03); C333(1.20) | LDD1561 | [13] |
| LDCM0323 | AC6 | HEK-293T | C203(1.05); C74(1.08); C323(0.98); C189(1.13) | LDD1562 | [13] |
| LDCM0324 | AC60 | HEK-293T | C203(1.05); C74(0.97); C323(0.97); C333(1.09) | LDD1563 | [13] |
| LDCM0325 | AC61 | HEK-293T | C203(0.98); C74(1.07); C323(0.99); C189(1.09) | LDD1564 | [13] |
| LDCM0326 | AC62 | HEK-293T | C203(1.09); C74(1.02); C323(1.01); C189(1.16) | LDD1565 | [13] |
| LDCM0327 | AC63 | HEK-293T | C203(0.96); C74(0.95); C323(1.01); C333(0.98) | LDD1566 | [13] |
| LDCM0328 | AC64 | HEK-293T | C203(1.07); C74(0.99); C323(0.98); C333(1.01) | LDD1567 | [13] |
| LDCM0334 | AC7 | HEK-293T | C203(0.90); C74(0.96); C323(1.03); C333(1.07) | LDD1568 | [13] |
| LDCM0345 | AC8 | HEK-293T | C203(1.14); C74(1.02); C323(1.02); C333(1.07) | LDD1569 | [13] |
| LDCM0248 | AKOS034007472 | HEK-293T | C203(0.95); C74(1.00); C323(0.97); C189(1.04) | LDD1511 | [13] |
| LDCM0356 | AKOS034007680 | HEK-293T | C203(1.20); C74(1.12); C323(0.99); C333(1.03) | LDD1570 | [13] |
| LDCM0275 | AKOS034007705 | HEK-293T | C203(1.07); C74(1.12); C323(0.96); C333(0.99) | LDD1514 | [13] |
| LDCM0156 | Aniline | NCI-H1299 | 12.05 | LDD0403 | [1] |
| LDCM0630 | CCW28-3 | 231MFP | C361(1.09) | LDD2214 | [14] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [6] |
| LDCM0367 | CL1 | HEK-293T | C203(1.00); C74(1.08); C323(1.06); C78(0.95) | LDD1571 | [13] |
| LDCM0368 | CL10 | HEK-293T | C203(0.94); C74(1.16); C323(0.82); C189(1.49) | LDD1572 | [13] |
| LDCM0369 | CL100 | HEK-293T | C203(0.97); C74(1.03); C323(1.00); C333(0.95) | LDD1573 | [13] |
| LDCM0370 | CL101 | HEK-293T | C203(1.08); C74(1.15); C323(1.02); C78(1.18) | LDD1574 | [13] |
| LDCM0371 | CL102 | HEK-293T | C203(1.01); C74(1.16); C323(0.98); C333(1.04) | LDD1575 | [13] |
| LDCM0372 | CL103 | HEK-293T | C203(1.32); C74(1.10); C323(1.02); C333(1.01) | LDD1576 | [13] |
| LDCM0373 | CL104 | HEK-293T | C203(0.95); C74(1.00); C323(1.02); C333(1.16) | LDD1577 | [13] |
| LDCM0374 | CL105 | HEK-293T | C203(1.04); C74(1.06); C323(1.00); C78(1.11) | LDD1578 | [13] |
| LDCM0375 | CL106 | HEK-293T | C203(0.87); C74(1.11); C323(0.96); C333(0.84) | LDD1579 | [13] |
| LDCM0376 | CL107 | HEK-293T | C203(1.14); C74(1.00); C323(1.02); C333(1.03) | LDD1580 | [13] |
| LDCM0377 | CL108 | HEK-293T | C203(0.87); C74(1.01); C323(0.92); C333(1.03) | LDD1581 | [13] |
| LDCM0378 | CL109 | HEK-293T | C203(0.97); C74(1.11); C323(1.00); C78(1.12) | LDD1582 | [13] |
| LDCM0379 | CL11 | HEK-293T | C203(0.94); C74(1.14); C323(0.92); C333(0.97) | LDD1583 | [13] |
| LDCM0380 | CL110 | HEK-293T | C203(1.02); C74(1.10); C323(0.92); C333(0.87) | LDD1584 | [13] |
| LDCM0381 | CL111 | HEK-293T | C203(1.01); C74(1.04); C323(0.98); C333(1.01) | LDD1585 | [13] |
| LDCM0382 | CL112 | HEK-293T | C203(0.82); C74(1.01); C323(0.94); C333(1.03) | LDD1586 | [13] |
| LDCM0383 | CL113 | HEK-293T | C203(1.03); C74(0.99); C323(1.00); C78(1.04) | LDD1587 | [13] |
| LDCM0384 | CL114 | HEK-293T | C203(0.85); C74(2.10); C323(0.95); C333(0.83) | LDD1588 | [13] |
| LDCM0385 | CL115 | HEK-293T | C203(1.16); C74(1.04); C323(1.02); C333(0.94) | LDD1589 | [13] |
| LDCM0386 | CL116 | HEK-293T | C203(1.06); C74(1.13); C323(1.07); C333(1.03) | LDD1590 | [13] |
| LDCM0387 | CL117 | HEK-293T | C203(0.99); C74(1.08); C323(1.07); C78(1.22) | LDD1591 | [13] |
| LDCM0388 | CL118 | HEK-293T | C203(1.04); C74(1.07); C323(1.02); C333(1.05) | LDD1592 | [13] |
| LDCM0389 | CL119 | HEK-293T | C203(1.08); C74(1.10); C323(0.97); C333(1.11) | LDD1593 | [13] |
| LDCM0390 | CL12 | HEK-293T | C203(0.97); C74(1.29); C323(1.06); C333(1.03) | LDD1594 | [13] |
| LDCM0391 | CL120 | HEK-293T | C203(1.01); C74(1.07); C323(1.02); C333(1.02) | LDD1595 | [13] |
| LDCM0392 | CL121 | HEK-293T | C203(1.10); C74(1.05); C323(1.16); C78(1.03) | LDD1596 | [13] |
| LDCM0393 | CL122 | HEK-293T | C203(0.90); C74(1.14); C323(1.01); C333(1.00) | LDD1597 | [13] |
| LDCM0394 | CL123 | HEK-293T | C203(1.02); C74(1.10); C323(0.94); C333(0.98) | LDD1598 | [13] |
| LDCM0395 | CL124 | HEK-293T | C203(1.09); C74(1.02); C323(0.95); C333(0.97) | LDD1599 | [13] |
| LDCM0396 | CL125 | HEK-293T | C203(1.07); C74(0.95); C323(1.02); C78(1.18) | LDD1600 | [13] |
| LDCM0397 | CL126 | HEK-293T | C203(0.98); C74(0.98); C323(1.03); C333(1.05) | LDD1601 | [13] |
| LDCM0398 | CL127 | HEK-293T | C203(1.18); C74(1.22); C323(0.99); C333(0.81) | LDD1602 | [13] |
| LDCM0399 | CL128 | HEK-293T | C203(1.05); C74(1.07); C323(1.00); C333(0.91) | LDD1603 | [13] |
| LDCM0400 | CL13 | HEK-293T | C203(1.20); C74(1.01); C323(1.03); C78(1.15) | LDD1604 | [13] |
| LDCM0401 | CL14 | HEK-293T | C203(1.05); C74(0.99); C323(1.06); C333(1.05) | LDD1605 | [13] |
| LDCM0402 | CL15 | HEK-293T | C203(1.10); C74(1.31); C323(0.92); C333(0.93) | LDD1606 | [13] |
| LDCM0403 | CL16 | HEK-293T | C203(1.00); C74(1.04); C323(1.03); C333(1.07) | LDD1607 | [13] |
| LDCM0404 | CL17 | HEK-293T | C203(1.09); C74(1.56); C323(0.96); C333(1.09) | LDD1608 | [13] |
| LDCM0405 | CL18 | HEK-293T | C203(1.05); C74(1.17); C323(1.09); C333(1.00) | LDD1609 | [13] |
| LDCM0406 | CL19 | HEK-293T | C203(0.91); C74(1.15); C323(1.01); C333(1.12) | LDD1610 | [13] |
| LDCM0407 | CL2 | HEK-293T | C203(0.97); C74(1.13); C323(1.05); C333(1.04) | LDD1611 | [13] |
| LDCM0408 | CL20 | HEK-293T | C203(1.16); C74(1.02); C323(0.91); C333(1.19) | LDD1612 | [13] |
| LDCM0409 | CL21 | HEK-293T | C203(0.95); C74(0.59); C323(0.89); C189(1.25) | LDD1613 | [13] |
| LDCM0410 | CL22 | HEK-293T | C203(0.99); C74(1.20); C323(0.91); C189(1.23) | LDD1614 | [13] |
| LDCM0411 | CL23 | HEK-293T | C203(0.90); C74(1.08); C323(0.94); C333(1.03) | LDD1615 | [13] |
| LDCM0412 | CL24 | HEK-293T | C203(1.06); C74(1.23); C323(0.99); C333(1.06) | LDD1616 | [13] |
| LDCM0413 | CL25 | HEK-293T | C203(1.03); C74(1.06); C323(1.07); C78(1.32) | LDD1617 | [13] |
| LDCM0414 | CL26 | HEK-293T | C203(0.97); C74(0.99); C323(0.99); C333(1.10) | LDD1618 | [13] |
| LDCM0415 | CL27 | HEK-293T | C203(1.09); C74(1.11); C323(0.96); C333(1.14) | LDD1619 | [13] |
| LDCM0416 | CL28 | HEK-293T | C203(0.87); C74(1.11); C323(0.94); C333(1.09) | LDD1620 | [13] |
| LDCM0417 | CL29 | HEK-293T | C203(1.14); C74(1.06); C323(0.99); C333(0.97) | LDD1621 | [13] |
| LDCM0418 | CL3 | HEK-293T | C203(1.28); C74(1.04); C323(1.01); C333(1.14) | LDD1622 | [13] |
| LDCM0419 | CL30 | HEK-293T | C203(1.00); C74(1.06); C323(1.00); C333(1.17) | LDD1623 | [13] |
| LDCM0420 | CL31 | HEK-293T | C203(0.95); C74(1.09); C323(0.98); C333(1.02) | LDD1624 | [13] |
| LDCM0421 | CL32 | HEK-293T | C203(1.05); C74(1.05); C323(0.94); C333(1.02) | LDD1625 | [13] |
| LDCM0422 | CL33 | HEK-293T | C203(0.81); C74(1.04); C323(0.88); C189(1.12) | LDD1626 | [13] |
| LDCM0423 | CL34 | HEK-293T | C203(0.86); C74(1.11); C323(0.89); C189(1.50) | LDD1627 | [13] |
| LDCM0424 | CL35 | HEK-293T | C203(0.87); C74(1.08); C323(0.85); C333(1.03) | LDD1628 | [13] |
| LDCM0425 | CL36 | HEK-293T | C203(1.00); C74(1.45); C323(0.81); C333(0.94) | LDD1629 | [13] |
| LDCM0426 | CL37 | HEK-293T | C203(0.98); C74(1.03); C323(1.12); C78(1.13) | LDD1630 | [13] |
| LDCM0428 | CL39 | HEK-293T | C203(1.13); C74(0.92); C323(0.89); C333(1.02) | LDD1632 | [13] |
| LDCM0429 | CL4 | HEK-293T | C203(0.88); C74(0.91); C323(1.03); C333(1.01) | LDD1633 | [13] |
| LDCM0430 | CL40 | HEK-293T | C203(0.91); C74(1.03); C323(0.95); C333(1.09) | LDD1634 | [13] |
| LDCM0431 | CL41 | HEK-293T | C203(0.98); C74(1.11); C323(0.89); C333(0.83) | LDD1635 | [13] |
| LDCM0432 | CL42 | HEK-293T | C203(0.93); C74(1.08); C323(1.01); C333(1.06) | LDD1636 | [13] |
| LDCM0433 | CL43 | HEK-293T | C203(0.84); C74(1.09); C323(1.06); C333(1.23) | LDD1637 | [13] |
| LDCM0434 | CL44 | HEK-293T | C203(0.97); C74(1.07); C323(1.02); C333(1.07) | LDD1638 | [13] |
| LDCM0435 | CL45 | HEK-293T | C203(0.92); C74(0.95); C323(0.89); C189(1.13) | LDD1639 | [13] |
| LDCM0436 | CL46 | HEK-293T | C203(0.87); C74(1.08); C323(0.94); C189(1.04) | LDD1640 | [13] |
| LDCM0437 | CL47 | HEK-293T | C203(0.88); C74(1.52); C323(0.85); C333(0.95) | LDD1641 | [13] |
| LDCM0438 | CL48 | HEK-293T | C203(0.96); C74(1.32); C323(0.94); C333(1.06) | LDD1642 | [13] |
| LDCM0439 | CL49 | HEK-293T | C203(1.11); C74(1.02); C323(1.06); C78(1.09) | LDD1643 | [13] |
| LDCM0440 | CL5 | HEK-293T | C203(1.07); C74(1.03); C323(0.88); C333(1.06) | LDD1644 | [13] |
| LDCM0441 | CL50 | HEK-293T | C203(0.97); C74(1.06); C323(1.01); C333(0.96) | LDD1645 | [13] |
| LDCM0443 | CL52 | HEK-293T | C203(0.92); C74(0.96); C323(0.97); C333(1.03) | LDD1646 | [13] |
| LDCM0444 | CL53 | HEK-293T | C203(1.13); C74(1.16); C323(0.88); C333(0.90) | LDD1647 | [13] |
| LDCM0445 | CL54 | HEK-293T | C203(0.99); C74(1.13); C323(0.95); C333(1.02) | LDD1648 | [13] |
| LDCM0446 | CL55 | HEK-293T | C203(0.94); C74(1.13); C323(0.94); C333(1.09) | LDD1649 | [13] |
| LDCM0447 | CL56 | HEK-293T | C203(1.16); C74(1.08); C323(0.97); C333(1.01) | LDD1650 | [13] |
| LDCM0448 | CL57 | HEK-293T | C203(0.99); C74(0.89); C323(0.82); C189(1.20) | LDD1651 | [13] |
| LDCM0449 | CL58 | HEK-293T | C203(0.98); C74(1.30); C323(0.85); C189(1.13) | LDD1652 | [13] |
| LDCM0450 | CL59 | HEK-293T | C203(0.82); C74(1.14); C323(0.85); C333(1.07) | LDD1653 | [13] |
| LDCM0451 | CL6 | HEK-293T | C203(1.01); C74(1.25); C323(0.91); C333(0.91) | LDD1654 | [13] |
| LDCM0452 | CL60 | HEK-293T | C203(1.08); C74(1.38); C323(0.93); C333(1.12) | LDD1655 | [13] |
| LDCM0453 | CL61 | HEK-293T | C203(1.02); C74(0.89); C323(1.17); C78(1.07) | LDD1656 | [13] |
| LDCM0454 | CL62 | HEK-293T | C203(0.85); C74(1.08); C323(1.13); C333(1.08) | LDD1657 | [13] |
| LDCM0455 | CL63 | HEK-293T | C203(1.19); C74(1.12); C323(1.02); C333(1.19) | LDD1658 | [13] |
| LDCM0456 | CL64 | HEK-293T | C203(0.89); C74(0.98); C323(0.94); C333(1.00) | LDD1659 | [13] |
| LDCM0457 | CL65 | HEK-293T | C203(1.15); C74(1.17); C323(0.98); C333(1.06) | LDD1660 | [13] |
| LDCM0458 | CL66 | HEK-293T | C203(0.90); C74(1.50); C323(0.81); C333(0.87) | LDD1661 | [13] |
| LDCM0459 | CL67 | HEK-293T | C203(0.93); C74(1.07); C323(1.00); C333(0.98) | LDD1662 | [13] |
| LDCM0460 | CL68 | HEK-293T | C203(1.05); C74(1.12); C323(1.03); C333(1.13) | LDD1663 | [13] |
| LDCM0461 | CL69 | HEK-293T | C203(0.88); C74(0.99); C323(0.90); C189(1.18) | LDD1664 | [13] |
| LDCM0462 | CL7 | HEK-293T | C203(0.88); C74(1.09); C323(1.06); C333(1.22) | LDD1665 | [13] |
| LDCM0463 | CL70 | HEK-293T | C203(0.93); C74(1.26); C323(0.94); C189(1.26) | LDD1666 | [13] |
| LDCM0464 | CL71 | HEK-293T | C203(0.82); C74(1.14); C323(0.91); C333(1.08) | LDD1667 | [13] |
| LDCM0465 | CL72 | HEK-293T | C203(1.03); C74(1.25); C323(0.95); C333(1.07) | LDD1668 | [13] |
| LDCM0466 | CL73 | HEK-293T | C203(1.12); C74(1.07); C323(1.03); C78(1.20) | LDD1669 | [13] |
| LDCM0467 | CL74 | HEK-293T | C203(0.88); C74(1.00); C323(0.98); C333(1.09) | LDD1670 | [13] |
| LDCM0469 | CL76 | HEK-293T | C203(1.00); C74(1.11); C323(1.00); C333(1.09) | LDD1672 | [13] |
| LDCM0470 | CL77 | HEK-293T | C203(1.12); C74(1.17); C323(0.86); C333(1.05) | LDD1673 | [13] |
| LDCM0471 | CL78 | HEK-293T | C203(0.96); C74(1.09); C323(1.03); C333(1.07) | LDD1674 | [13] |
| LDCM0472 | CL79 | HEK-293T | C203(0.97); C74(1.14); C323(1.00); C333(1.14) | LDD1675 | [13] |
| LDCM0473 | CL8 | HEK-293T | C203(1.11); C74(0.96); C323(0.94); C333(1.05) | LDD1676 | [13] |
| LDCM0474 | CL80 | HEK-293T | C203(1.07); C74(1.12); C323(1.01); C333(1.07) | LDD1677 | [13] |
| LDCM0475 | CL81 | HEK-293T | C203(0.84); C74(1.11); C323(0.90); C189(1.35) | LDD1678 | [13] |
| LDCM0476 | CL82 | HEK-293T | C203(0.96); C74(1.31); C323(1.03); C189(1.36) | LDD1679 | [13] |
| LDCM0477 | CL83 | HEK-293T | C203(0.87); C74(1.42); C323(0.80); C333(0.87) | LDD1680 | [13] |
| LDCM0478 | CL84 | HEK-293T | C203(1.06); C74(1.08); C323(0.89); C333(1.02) | LDD1681 | [13] |
| LDCM0479 | CL85 | HEK-293T | C203(1.12); C74(0.94); C323(1.08); C78(1.13) | LDD1682 | [13] |
| LDCM0480 | CL86 | HEK-293T | C203(1.06); C74(0.99); C323(1.08); C333(1.05) | LDD1683 | [13] |
| LDCM0481 | CL87 | HEK-293T | C203(1.36); C74(0.95); C323(0.94); C333(0.96) | LDD1684 | [13] |
| LDCM0482 | CL88 | HEK-293T | C203(1.02); C74(1.11); C323(1.03); C333(1.02) | LDD1685 | [13] |
| LDCM0483 | CL89 | HEK-293T | C203(1.05); C74(1.08); C323(0.98); C333(1.06) | LDD1686 | [13] |
| LDCM0484 | CL9 | HEK-293T | C203(0.80); C74(1.03); C323(0.91); C189(1.37) | LDD1687 | [13] |
| LDCM0485 | CL90 | HEK-293T | C203(1.00); C74(1.78); C323(0.77); C333(0.89) | LDD1688 | [13] |
| LDCM0486 | CL91 | HEK-293T | C203(0.94); C74(1.20); C323(1.09); C333(1.18) | LDD1689 | [13] |
| LDCM0487 | CL92 | HEK-293T | C203(1.05); C74(1.05); C323(1.03); C333(1.10) | LDD1690 | [13] |
| LDCM0488 | CL93 | HEK-293T | C203(0.86); C74(1.05); C323(0.94); C189(1.32) | LDD1691 | [13] |
| LDCM0489 | CL94 | HEK-293T | C203(0.99); C74(1.19); C323(0.90); C189(1.35) | LDD1692 | [13] |
| LDCM0490 | CL95 | HEK-293T | C203(0.96); C74(0.96); C323(0.84); C333(0.79) | LDD1693 | [13] |
| LDCM0491 | CL96 | HEK-293T | C203(1.08); C74(1.21); C323(0.92); C333(0.99) | LDD1694 | [13] |
| LDCM0492 | CL97 | HEK-293T | C203(1.02); C74(1.14); C323(0.96); C78(1.10) | LDD1695 | [13] |
| LDCM0493 | CL98 | HEK-293T | C203(1.03); C74(1.09); C323(1.01); C333(0.90) | LDD1696 | [13] |
| LDCM0494 | CL99 | HEK-293T | C203(1.14); C74(0.95); C323(0.92); C333(0.97) | LDD1697 | [13] |
| LDCM0634 | CY-0357 | Hep-G2 | C189(0.82) | LDD2228 | [8] |
| LDCM0027 | Dopamine | HEK-293T | 11.07 | LDD0179 | [4] |
| LDCM0495 | E2913 | HEK-293T | C203(1.22); C74(1.08); C323(0.96); C333(1.08) | LDD1698 | [13] |
| LDCM0625 | F8 | Ramos | C74(1.02); C333(1.20); C189(1.01) | LDD2187 | [15] |
| LDCM0572 | Fragment10 | Ramos | C74(0.61); C333(0.98); C189(1.02) | LDD2189 | [15] |
| LDCM0573 | Fragment11 | Ramos | C74(0.32); C189(0.98) | LDD2190 | [15] |
| LDCM0574 | Fragment12 | Ramos | C74(0.47); C333(0.72); C189(0.66) | LDD2191 | [15] |
| LDCM0575 | Fragment13 | Ramos | C74(0.55); C189(1.06) | LDD2192 | [15] |
| LDCM0576 | Fragment14 | Ramos | C74(1.12); C333(0.88); C189(0.83) | LDD2193 | [15] |
| LDCM0579 | Fragment20 | Ramos | C74(0.35); C333(0.67); C189(0.74) | LDD2194 | [15] |
| LDCM0580 | Fragment21 | Ramos | C74(0.51); C333(1.12); C189(0.73) | LDD2195 | [15] |
| LDCM0582 | Fragment23 | Ramos | C74(0.54); C333(1.16); C189(0.67) | LDD2196 | [15] |
| LDCM0578 | Fragment27 | Ramos | C74(0.70); C189(0.95) | LDD2197 | [15] |
| LDCM0586 | Fragment28 | Ramos | C74(1.54); C189(0.71) | LDD2198 | [15] |
| LDCM0588 | Fragment30 | Ramos | C74(0.63); C333(0.73); C189(0.72) | LDD2199 | [15] |
| LDCM0589 | Fragment31 | Ramos | C74(0.73); C189(0.82) | LDD2200 | [15] |
| LDCM0590 | Fragment32 | Ramos | C74(0.72); C333(0.68); C189(0.72) | LDD2201 | [15] |
| LDCM0468 | Fragment33 | HEK-293T | C203(1.16); C74(1.01); C323(0.95); C333(0.91) | LDD1671 | [13] |
| LDCM0596 | Fragment38 | Ramos | C74(0.33); C333(0.80); C189(0.86) | LDD2203 | [15] |
| LDCM0566 | Fragment4 | Ramos | C74(1.24); C333(0.44); C189(0.53) | LDD2184 | [15] |
| LDCM0427 | Fragment51 | HEK-293T | C203(0.90); C74(1.02); C323(1.08); C333(1.13) | LDD1631 | [13] |
| LDCM0610 | Fragment52 | Ramos | C74(0.23); C333(1.25); C189(0.84) | LDD2204 | [15] |
| LDCM0614 | Fragment56 | Ramos | C74(0.22); C333(2.21); C189(0.92) | LDD2205 | [15] |
| LDCM0569 | Fragment7 | Ramos | C74(1.17); C333(0.53); C189(0.79) | LDD2186 | [15] |
| LDCM0571 | Fragment9 | Ramos | C74(0.34); C333(0.81); C189(0.83) | LDD2188 | [15] |
| LDCM0022 | KB02 | HEK-293T | C203(1.00); C189(1.01); C333(0.93); C74(0.83) | LDD1492 | [13] |
| LDCM0023 | KB03 | HEK-293T | C203(1.06); C189(1.02); C333(0.87); C74(0.91) | LDD1497 | [13] |
| LDCM0024 | KB05 | IGR37 | C277(1.34) | LDD3314 | [3] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C74(0.74); C189(0.25) | LDD2207 | [16] |
| LDCM0131 | RA190 | MM1.R | C189(1.39) | LDD0304 | [17] |
| LDCM0113 | W17 | Hep-G2 | C189(9.46) | LDD0240 | [2] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Gamma-aminobutyric acid receptor-associated protein (GABARAP) | ATG8 family | O95166 | |||
Other
References
