Details of the Target
General Information of Target
| Target ID | LDTP13162 | |||||
|---|---|---|---|---|---|---|
| Target Name | Mortality factor 4-like protein 1 (MORF4L1) | |||||
| Gene Name | MORF4L1 | |||||
| Gene ID | 10933 | |||||
| Synonyms |
MRG15; Mortality factor 4-like protein 1; MORF-related gene 15 protein; MRG15; Protein MSL3-1; Transcription factor-like protein MRG15 |
|||||
| 3D Structure | ||||||
| Sequence |
MNSGAMRIHSKGHFQGGIQVKNEKNRPSLKSLKTDNRPEKSKCKPLWGKVFYLDLPSVTI
SEKLQKDIKDLGGRVEEFLSKDISYLISNKKEAKFAQTLGRISPVPSPESAYTAETTSPH PSHDGSSFKSPDTVCLSRGKLLVEKAIKDHDFIPSNSILSNALSWGVKILHIDDIRYYIE QKKKELYLLKKSSTSVRDGGKRVGSGAQKTRTGRLKKPFVKVEDMSQLYRPFYLQLTNMP FINYSIQKPCSPFDVDKPSSMQKQTQVKLRIQTDGDKYGGTSIQLQLKEKKKKGYCECCL QKYEDLETHLLSEQHRNFAQSNQYQVVDDIVSKLVFDFVEYEKDTPKKKRIKYSVGSLSP VSASVLKKTEQKEKVELQHISQKDCQEDDTTVKEQNFLYKETQETEKKLLFISEPIPHPS NELRGLNEKMSNKCSMLSTAEDDIRQNFTQLPLHKNKQECILDISEHTLSENDLEELRVD HYKCNIQASVHVSDFSTDNSGSQPKQKSDTVLFPAKDLKEKDLHSIFTHDSGLITINSSQ EHLTVQAKAPFHTPPEEPNECDFKNMDSLPSGKIHRKVKIILGRNRKENLEPNAEFDKRT EFITQEENRICSSPVQSLLDLFQTSEEKSEFLGFTSYTEKSGICNVLDIWEEENSDNLLT AFFSSPSTSTFTGF |
|||||
| Target Bioclass |
Other
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Component of the NuA4 histone acetyltransferase (HAT) complex which is involved in transcriptional activation of select genes principally by acetylation of nucleosomal histones H4 and H2A. This modification may both alter nucleosome - DNA interactions and promote interaction of the modified histones with other proteins which positively regulate transcription. This complex may be required for the activation of transcriptional programs associated with oncogene and proto-oncogene mediated growth induction, tumor suppressor mediated growth arrest and replicative senescence, apoptosis, and DNA repair. The NuA4 complex ATPase and helicase activities seem to be, at least in part, contributed by the association of RUVBL1 and RUVBL2 with EP400. NuA4 may also play a direct role in DNA repair when directly recruited to sites of DNA damage. As part of the SIN3B complex represses transcription and counteracts the histone acetyltransferase activity of EP300 through the recognition H3K27ac marks by PHF12 and the activity of the histone deacetylase HDAC2. SIN3B complex is recruited downstream of the constitutively active genes transcriptional start sites through interaction with histones and mitigates histone acetylation and RNA polymerase II progression within transcribed regions contributing to the regulation of transcription. Required for homologous recombination repair (HRR) and resistance to mitomycin C (MMC). Involved in the localization of PALB2, BRCA2 and RAD51, but not BRCA1, to DNA-damage foci.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K111(6.00); K127(10.00); K218(8.86); K240(7.14) | LDD0277 | [1] | |
|
Probe 1 Probe Info |
 |
Y104(4.82); Y123(47.81); Y239(11.36) | LDD3495 | [2] | |
|
BTD Probe Info |
 |
C19(1.27) | LDD2094 | [3] | |
|
HHS-475 Probe Info |
 |
Y239(0.89) | LDD0264 | [4] | |
|
DBIA Probe Info |
 |
C19(0.92) | LDD0593 | [5] | |
|
N1 Probe Info |
 |
K111(0.00); R113(0.00) | LDD0245 | [6] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [7] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [7] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [7] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [8] | |
|
SF Probe Info |
 |
Y239(0.00); K240(0.00) | LDD0028 | [9] | |
|
Ox-W18 Probe Info |
 |
W206(0.00); W211(0.00) | LDD2175 | [10] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [11] | |
|
HHS-465 Probe Info |
 |
N.A. | LDD2240 | [12] | |
|
HHS-482 Probe Info |
 |
Y123(1.09) | LDD2239 | [13] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C106 Probe Info |
 |
18.00 | LDD1793 | [14] | |
|
C193 Probe Info |
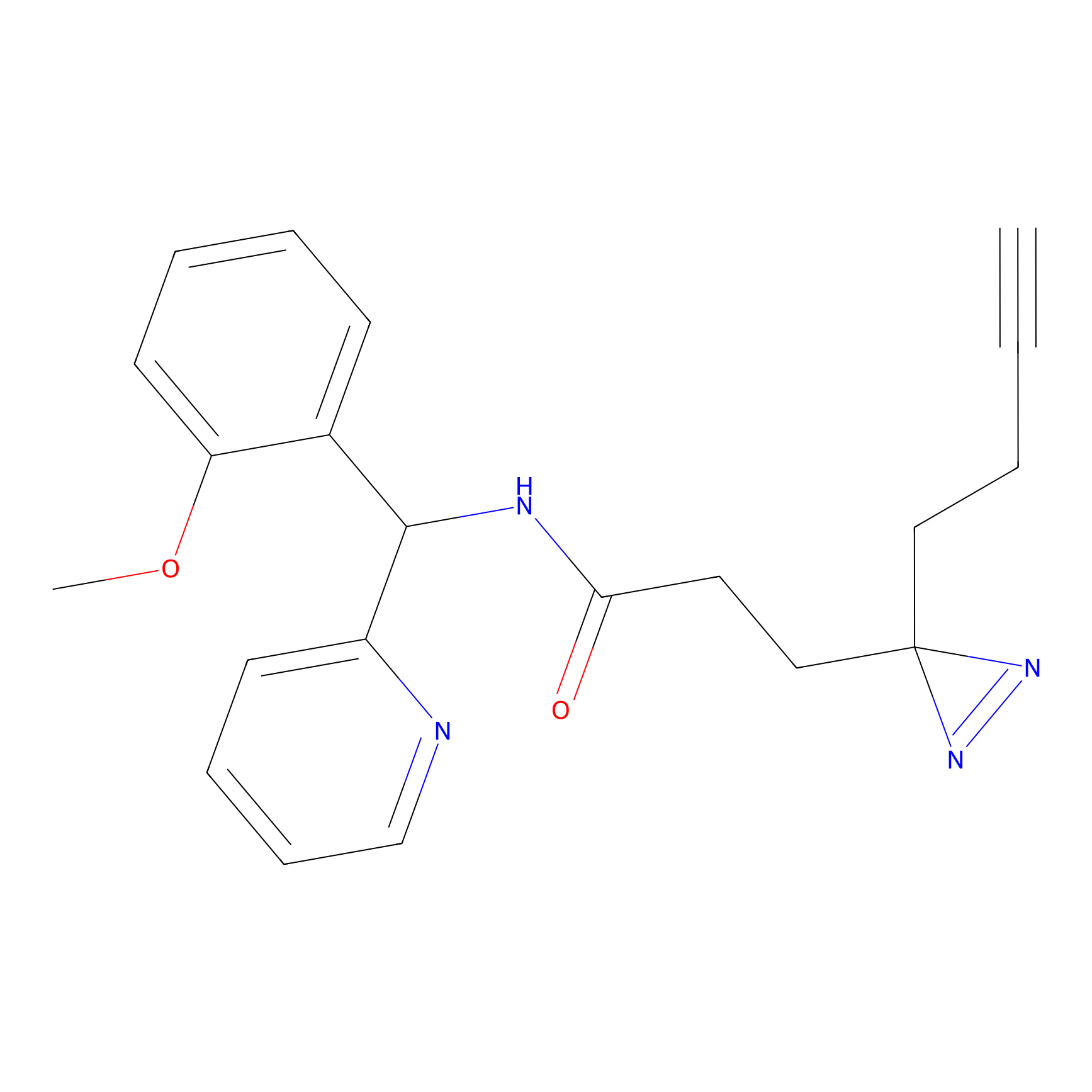 |
6.23 | LDD1869 | [14] | |
|
C350 Probe Info |
 |
24.93 | LDD2011 | [14] | |
|
C431 Probe Info |
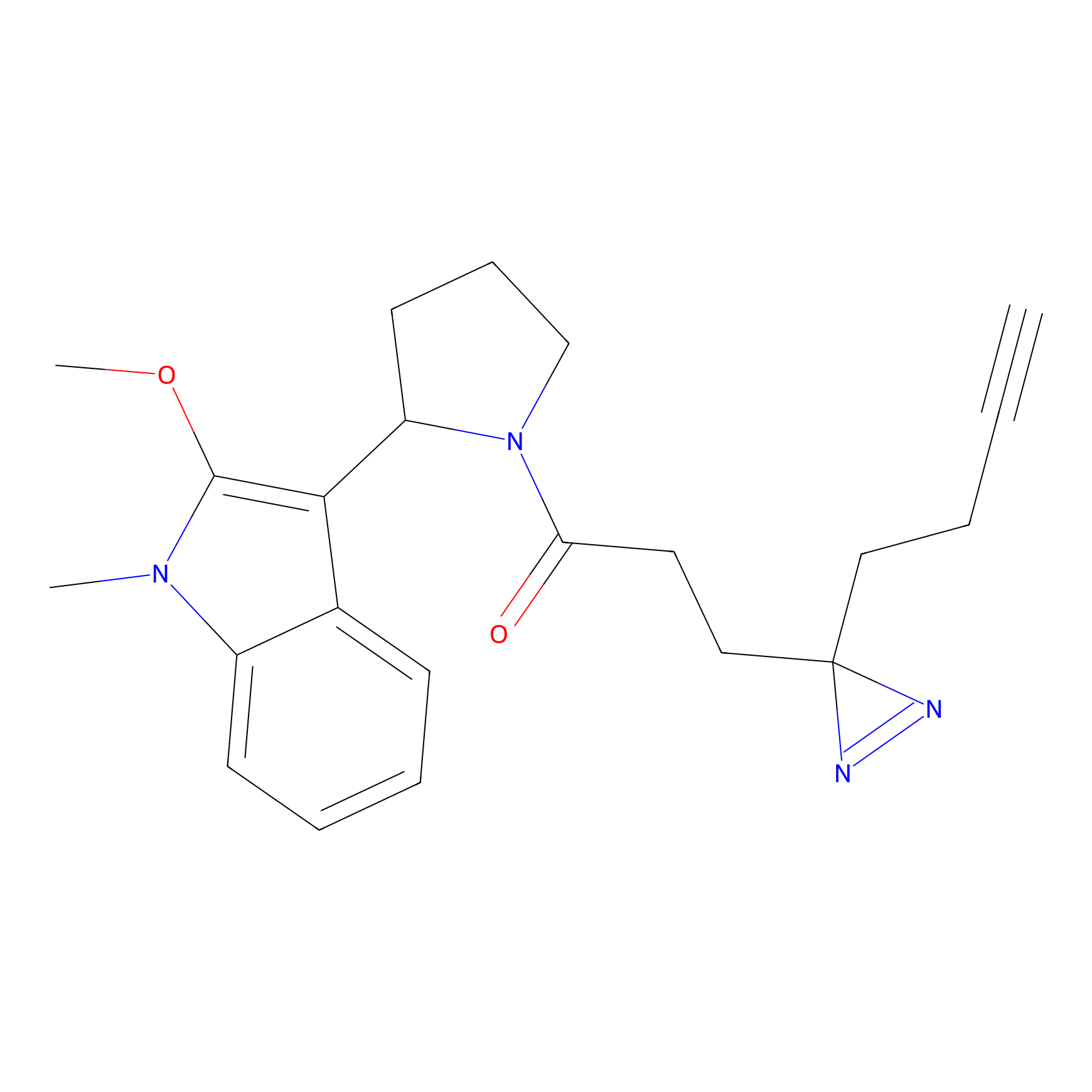 |
14.52 | LDD2086 | [14] | |
|
FFF probe11 Probe Info |
 |
20.00 | LDD0471 | [15] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [15] | |
|
FFF probe14 Probe Info |
 |
12.09 | LDD0477 | [15] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HEK-293T | C19(1.04) | LDD1507 | [16] |
| LDCM0215 | AC10 | HEK-293T | C19(0.77) | LDD1508 | [16] |
| LDCM0226 | AC11 | HEK-293T | C19(0.91) | LDD1509 | [16] |
| LDCM0237 | AC12 | HEK-293T | C19(0.95) | LDD1510 | [16] |
| LDCM0276 | AC17 | HCT 116 | C19(0.92) | LDD0593 | [5] |
| LDCM0277 | AC18 | HCT 116 | C19(0.45) | LDD0594 | [5] |
| LDCM0278 | AC19 | HCT 116 | C19(0.88) | LDD0595 | [5] |
| LDCM0279 | AC2 | HEK-293T | C19(1.08) | LDD1518 | [16] |
| LDCM0280 | AC20 | HCT 116 | C19(0.94) | LDD0597 | [5] |
| LDCM0281 | AC21 | HCT 116 | C19(0.78) | LDD0598 | [5] |
| LDCM0282 | AC22 | HCT 116 | C19(0.84) | LDD0599 | [5] |
| LDCM0283 | AC23 | HCT 116 | C19(0.74) | LDD0600 | [5] |
| LDCM0284 | AC24 | HCT 116 | C19(0.90) | LDD0601 | [5] |
| LDCM0285 | AC25 | HEK-293T | C19(0.91) | LDD1524 | [16] |
| LDCM0286 | AC26 | HEK-293T | C19(0.87) | LDD1525 | [16] |
| LDCM0287 | AC27 | HEK-293T | C19(1.03) | LDD1526 | [16] |
| LDCM0288 | AC28 | HEK-293T | C19(0.92) | LDD1527 | [16] |
| LDCM0289 | AC29 | HEK-293T | C19(1.06) | LDD1528 | [16] |
| LDCM0290 | AC3 | HEK-293T | C19(0.92) | LDD1529 | [16] |
| LDCM0293 | AC32 | HEK-293T | C19(1.27) | LDD1532 | [16] |
| LDCM0294 | AC33 | HEK-293T | C19(1.28) | LDD1533 | [16] |
| LDCM0295 | AC34 | HEK-293T | C19(0.85) | LDD1534 | [16] |
| LDCM0296 | AC35 | HCT 116 | C19(1.56) | LDD0613 | [5] |
| LDCM0297 | AC36 | HCT 116 | C19(0.98) | LDD0614 | [5] |
| LDCM0298 | AC37 | HCT 116 | C19(1.13) | LDD0615 | [5] |
| LDCM0299 | AC38 | HCT 116 | C19(1.15) | LDD0616 | [5] |
| LDCM0300 | AC39 | HCT 116 | C19(1.15) | LDD0617 | [5] |
| LDCM0301 | AC4 | HEK-293T | C19(0.97) | LDD1540 | [16] |
| LDCM0302 | AC40 | HCT 116 | C19(0.76) | LDD0619 | [5] |
| LDCM0303 | AC41 | HCT 116 | C19(1.20) | LDD0620 | [5] |
| LDCM0304 | AC42 | HCT 116 | C19(0.84) | LDD0621 | [5] |
| LDCM0305 | AC43 | HCT 116 | C19(1.23) | LDD0622 | [5] |
| LDCM0306 | AC44 | HCT 116 | C19(0.94) | LDD0623 | [5] |
| LDCM0307 | AC45 | HCT 116 | C19(0.69) | LDD0624 | [5] |
| LDCM0310 | AC48 | HEK-293T | C19(0.97) | LDD1549 | [16] |
| LDCM0311 | AC49 | HEK-293T | C19(0.97) | LDD1550 | [16] |
| LDCM0312 | AC5 | HEK-293T | C19(1.02) | LDD1551 | [16] |
| LDCM0313 | AC50 | HEK-293T | C19(0.87) | LDD1552 | [16] |
| LDCM0314 | AC51 | HEK-293T | C19(1.01) | LDD1553 | [16] |
| LDCM0315 | AC52 | HEK-293T | C19(0.99) | LDD1554 | [16] |
| LDCM0316 | AC53 | HEK-293T | C19(0.95) | LDD1555 | [16] |
| LDCM0319 | AC56 | HEK-293T | C19(1.14) | LDD1558 | [16] |
| LDCM0320 | AC57 | HEK-293T | C19(1.08) | LDD1559 | [16] |
| LDCM0321 | AC58 | HEK-293T | C19(0.90) | LDD1560 | [16] |
| LDCM0322 | AC59 | HEK-293T | C19(0.91) | LDD1561 | [16] |
| LDCM0324 | AC60 | HEK-293T | C19(0.90) | LDD1563 | [16] |
| LDCM0325 | AC61 | HEK-293T | C19(0.86) | LDD1564 | [16] |
| LDCM0328 | AC64 | HEK-293T | C19(1.02) | LDD1567 | [16] |
| LDCM0345 | AC8 | HEK-293T | C19(1.04) | LDD1569 | [16] |
| LDCM0248 | AKOS034007472 | HEK-293T | C19(0.99) | LDD1511 | [16] |
| LDCM0356 | AKOS034007680 | HEK-293T | C19(1.18) | LDD1570 | [16] |
| LDCM0275 | AKOS034007705 | HEK-293T | C19(0.95) | LDD1514 | [16] |
| LDCM0367 | CL1 | HEK-293T | C19(0.91) | LDD1571 | [16] |
| LDCM0369 | CL100 | HEK-293T | C19(1.13) | LDD1573 | [16] |
| LDCM0370 | CL101 | HEK-293T | C19(0.77) | LDD1574 | [16] |
| LDCM0371 | CL102 | HEK-293T | C19(0.88) | LDD1575 | [16] |
| LDCM0372 | CL103 | HEK-293T | C19(0.92) | LDD1576 | [16] |
| LDCM0373 | CL104 | HEK-293T | C19(1.09) | LDD1577 | [16] |
| LDCM0374 | CL105 | HCT 116 | C19(0.67) | LDD0691 | [5] |
| LDCM0375 | CL106 | HCT 116 | C19(0.49) | LDD0692 | [5] |
| LDCM0376 | CL107 | HCT 116 | C19(0.48) | LDD0693 | [5] |
| LDCM0377 | CL108 | HCT 116 | C19(0.41) | LDD0694 | [5] |
| LDCM0378 | CL109 | HCT 116 | C19(0.66) | LDD0695 | [5] |
| LDCM0380 | CL110 | HCT 116 | C19(0.66) | LDD0697 | [5] |
| LDCM0381 | CL111 | HCT 116 | C19(0.69) | LDD0698 | [5] |
| LDCM0382 | CL112 | HEK-293T | C19(1.29) | LDD1586 | [16] |
| LDCM0383 | CL113 | HEK-293T | C19(0.89) | LDD1587 | [16] |
| LDCM0384 | CL114 | HEK-293T | C19(0.99) | LDD1588 | [16] |
| LDCM0385 | CL115 | HEK-293T | C19(1.08) | LDD1589 | [16] |
| LDCM0386 | CL116 | HEK-293T | C19(0.99) | LDD1590 | [16] |
| LDCM0387 | CL117 | HCT 116 | C19(0.55) | LDD0704 | [5] |
| LDCM0388 | CL118 | HCT 116 | C19(1.16) | LDD0705 | [5] |
| LDCM0389 | CL119 | HCT 116 | C19(0.77) | LDD0706 | [5] |
| LDCM0390 | CL12 | HEK-293T | C19(0.94) | LDD1594 | [16] |
| LDCM0391 | CL120 | HCT 116 | C19(1.06) | LDD0708 | [5] |
| LDCM0392 | CL121 | HEK-293T | C19(0.80) | LDD1596 | [16] |
| LDCM0393 | CL122 | HEK-293T | C19(0.94) | LDD1597 | [16] |
| LDCM0394 | CL123 | HEK-293T | C19(0.99) | LDD1598 | [16] |
| LDCM0395 | CL124 | HEK-293T | C19(0.98) | LDD1599 | [16] |
| LDCM0396 | CL125 | HEK-293T | C19(0.83) | LDD1600 | [16] |
| LDCM0397 | CL126 | HEK-293T | C19(0.95) | LDD1601 | [16] |
| LDCM0398 | CL127 | HEK-293T | C19(1.06) | LDD1602 | [16] |
| LDCM0399 | CL128 | HEK-293T | C19(1.05) | LDD1603 | [16] |
| LDCM0400 | CL13 | HEK-293T | C19(0.94) | LDD1604 | [16] |
| LDCM0401 | CL14 | HEK-293T | C19(0.87) | LDD1605 | [16] |
| LDCM0402 | CL15 | HEK-293T | C19(1.01) | LDD1606 | [16] |
| LDCM0403 | CL16 | HCT 116 | C19(0.80) | LDD0720 | [5] |
| LDCM0404 | CL17 | HCT 116 | C19(0.80) | LDD0721 | [5] |
| LDCM0405 | CL18 | HCT 116 | C19(0.91) | LDD0722 | [5] |
| LDCM0406 | CL19 | HCT 116 | C19(0.86) | LDD0723 | [5] |
| LDCM0407 | CL2 | HEK-293T | C19(0.90) | LDD1611 | [16] |
| LDCM0408 | CL20 | HCT 116 | C19(0.66) | LDD0725 | [5] |
| LDCM0409 | CL21 | HCT 116 | C19(0.65) | LDD0726 | [5] |
| LDCM0410 | CL22 | HCT 116 | C19(0.45) | LDD0727 | [5] |
| LDCM0411 | CL23 | HCT 116 | C19(0.69) | LDD0728 | [5] |
| LDCM0412 | CL24 | HCT 116 | C19(0.51) | LDD0729 | [5] |
| LDCM0413 | CL25 | HCT 116 | C19(0.51) | LDD0730 | [5] |
| LDCM0414 | CL26 | HCT 116 | C19(0.58) | LDD0731 | [5] |
| LDCM0415 | CL27 | HCT 116 | C19(0.78) | LDD0732 | [5] |
| LDCM0416 | CL28 | HCT 116 | C19(0.78) | LDD0733 | [5] |
| LDCM0417 | CL29 | HCT 116 | C19(0.76) | LDD0734 | [5] |
| LDCM0418 | CL3 | HEK-293T | C19(1.05) | LDD1622 | [16] |
| LDCM0419 | CL30 | HCT 116 | C19(0.85) | LDD0736 | [5] |
| LDCM0420 | CL31 | HCT 116 | C19(0.73) | LDD0737 | [5] |
| LDCM0421 | CL32 | HCT 116 | C19(0.79) | LDD0738 | [5] |
| LDCM0422 | CL33 | HCT 116 | C19(0.85) | LDD0739 | [5] |
| LDCM0423 | CL34 | HCT 116 | C19(0.54) | LDD0740 | [5] |
| LDCM0424 | CL35 | HCT 116 | C19(0.47) | LDD0741 | [5] |
| LDCM0425 | CL36 | HCT 116 | C19(0.55) | LDD0742 | [5] |
| LDCM0426 | CL37 | HCT 116 | C19(0.44) | LDD0743 | [5] |
| LDCM0428 | CL39 | HCT 116 | C19(0.48) | LDD0745 | [5] |
| LDCM0429 | CL4 | HEK-293T | C19(1.20) | LDD1633 | [16] |
| LDCM0430 | CL40 | HCT 116 | C19(0.48) | LDD0747 | [5] |
| LDCM0431 | CL41 | HCT 116 | C19(0.62) | LDD0748 | [5] |
| LDCM0432 | CL42 | HCT 116 | C19(0.38) | LDD0749 | [5] |
| LDCM0433 | CL43 | HCT 116 | C19(0.47) | LDD0750 | [5] |
| LDCM0434 | CL44 | HCT 116 | C19(0.62) | LDD0751 | [5] |
| LDCM0435 | CL45 | HCT 116 | C19(0.61) | LDD0752 | [5] |
| LDCM0438 | CL48 | HEK-293T | C19(0.91) | LDD1642 | [16] |
| LDCM0439 | CL49 | HEK-293T | C19(0.71) | LDD1643 | [16] |
| LDCM0440 | CL5 | HEK-293T | C19(1.05) | LDD1644 | [16] |
| LDCM0441 | CL50 | HEK-293T | C19(0.96) | LDD1645 | [16] |
| LDCM0443 | CL52 | HEK-293T | C19(1.02) | LDD1646 | [16] |
| LDCM0444 | CL53 | HEK-293T | C19(0.95) | LDD1647 | [16] |
| LDCM0445 | CL54 | HEK-293T | C19(0.71) | LDD1648 | [16] |
| LDCM0446 | CL55 | HEK-293T | C19(1.00) | LDD1649 | [16] |
| LDCM0447 | CL56 | HEK-293T | C19(0.88) | LDD1650 | [16] |
| LDCM0448 | CL57 | HEK-293T | C19(0.88) | LDD1651 | [16] |
| LDCM0451 | CL6 | HEK-293T | C19(0.75) | LDD1654 | [16] |
| LDCM0452 | CL60 | HEK-293T | C19(1.21) | LDD1655 | [16] |
| LDCM0453 | CL61 | HEK-293T | C19(0.89) | LDD1656 | [16] |
| LDCM0454 | CL62 | HEK-293T | C19(0.96) | LDD1657 | [16] |
| LDCM0455 | CL63 | HEK-293T | C19(0.95) | LDD1658 | [16] |
| LDCM0456 | CL64 | HEK-293T | C19(0.94) | LDD1659 | [16] |
| LDCM0457 | CL65 | HEK-293T | C19(1.02) | LDD1660 | [16] |
| LDCM0458 | CL66 | HEK-293T | C19(0.96) | LDD1661 | [16] |
| LDCM0459 | CL67 | HEK-293T | C19(1.01) | LDD1662 | [16] |
| LDCM0460 | CL68 | HEK-293T | C19(0.94) | LDD1663 | [16] |
| LDCM0461 | CL69 | HEK-293T | C19(0.93) | LDD1664 | [16] |
| LDCM0462 | CL7 | HEK-293T | C19(0.80) | LDD1665 | [16] |
| LDCM0465 | CL72 | HEK-293T | C19(1.08) | LDD1668 | [16] |
| LDCM0466 | CL73 | HEK-293T | C19(0.75) | LDD1669 | [16] |
| LDCM0467 | CL74 | HEK-293T | C19(1.03) | LDD1670 | [16] |
| LDCM0469 | CL76 | HEK-293T | C19(1.04) | LDD1672 | [16] |
| LDCM0470 | CL77 | HEK-293T | C19(0.88) | LDD1673 | [16] |
| LDCM0471 | CL78 | HEK-293T | C19(0.89) | LDD1674 | [16] |
| LDCM0472 | CL79 | HEK-293T | C19(0.86) | LDD1675 | [16] |
| LDCM0473 | CL8 | HEK-293T | C19(0.76) | LDD1676 | [16] |
| LDCM0474 | CL80 | HEK-293T | C19(0.93) | LDD1677 | [16] |
| LDCM0475 | CL81 | HEK-293T | C19(0.91) | LDD1678 | [16] |
| LDCM0478 | CL84 | HEK-293T | C19(1.07) | LDD1681 | [16] |
| LDCM0479 | CL85 | HEK-293T | C19(1.10) | LDD1682 | [16] |
| LDCM0480 | CL86 | HEK-293T | C19(0.95) | LDD1683 | [16] |
| LDCM0481 | CL87 | HEK-293T | C19(0.92) | LDD1684 | [16] |
| LDCM0482 | CL88 | HEK-293T | C19(1.13) | LDD1685 | [16] |
| LDCM0483 | CL89 | HEK-293T | C19(0.93) | LDD1686 | [16] |
| LDCM0484 | CL9 | HEK-293T | C19(1.00) | LDD1687 | [16] |
| LDCM0485 | CL90 | HEK-293T | C19(0.63) | LDD1688 | [16] |
| LDCM0486 | CL91 | HEK-293T | C19(1.07) | LDD1689 | [16] |
| LDCM0487 | CL92 | HEK-293T | C19(0.86) | LDD1690 | [16] |
| LDCM0488 | CL93 | HEK-293T | C19(0.97) | LDD1691 | [16] |
| LDCM0491 | CL96 | HEK-293T | C19(1.01) | LDD1694 | [16] |
| LDCM0492 | CL97 | HEK-293T | C19(0.74) | LDD1695 | [16] |
| LDCM0493 | CL98 | HEK-293T | C19(0.96) | LDD1696 | [16] |
| LDCM0494 | CL99 | HEK-293T | C19(0.93) | LDD1697 | [16] |
| LDCM0495 | E2913 | HEK-293T | C19(1.16) | LDD1698 | [16] |
| LDCM0468 | Fragment33 | HEK-293T | C19(0.93) | LDD1671 | [16] |
| LDCM0427 | Fragment51 | HCT 116 | C19(0.41) | LDD0744 | [5] |
| LDCM0116 | HHS-0101 | DM93 | Y239(0.89) | LDD0264 | [4] |
| LDCM0117 | HHS-0201 | DM93 | Y239(0.49) | LDD0265 | [4] |
| LDCM0119 | HHS-0401 | DM93 | Y239(3.59) | LDD0267 | [4] |
| LDCM0120 | HHS-0701 | DM93 | Y239(2.09) | LDD0268 | [4] |
| LDCM0022 | KB02 | A2058 | C19(1.55) | LDD2253 | [17] |
| LDCM0023 | KB03 | A2058 | C19(2.04) | LDD2670 | [17] |
| LDCM0024 | KB05 | NCI-H3122 | C19(1.91) | LDD3360 | [17] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C19(1.27) | LDD2094 | [3] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C19(0.99) | LDD2098 | [3] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C19(0.68) | LDD2109 | [3] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C19(0.55) | LDD2114 | [3] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C19(0.61) | LDD2123 | [3] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C19(1.38) | LDD2136 | [3] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
Transcription factor
Other
References
