Details of the Target
General Information of Target
| Target ID | LDTP01246 | |||||
|---|---|---|---|---|---|---|
| Target Name | KH domain-containing, RNA-binding, signal transduction-associated protein 3 (KHDRBS3) | |||||
| Gene Name | KHDRBS3 | |||||
| Gene ID | 10656 | |||||
| Synonyms |
SALP; SLM2; KH domain-containing, RNA-binding, signal transduction-associated protein 3; RNA-binding protein T-Star; Sam68-like mammalian protein 2; SLM-2; Sam68-like phosphotyrosine protein |
|||||
| 3D Structure | ||||||
| Sequence |
MEEKYLPELMAEKDSLDPSFTHALRLVNQEIEKFQKGEGKDEEKYIDVVINKNMKLGQKV
LIPVKQFPKFNFVGKLLGPRGNSLKRLQEETLTKMSILGKGSMRDKAKEEELRKSGEAKY FHLNDDLHVLIEVFAPPAEAYARMGHALEEIKKFLIPDYNDEIRQAQLQELTYLNGGSEN ADVPVVRGKPTLRTRGVPAPAITRGRGGVTARPVGVVVPRGTPTPRGVLSTRGPVSRGRG LLTPRARGVPPTGYRPPPPPPTQETYGEYDYDDGYGTAYDEQSYDSYDNSYSTPAQSGAD YYDYGHGLSEETYDSYGQEEWTNSRHKAPSARTAKGVYRDQPYGRY |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
KHDRBS family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
RNA-binding protein that plays a role in the regulation of alternative splicing and influences mRNA splice site selection and exon inclusion. Binds preferentially to the 5'-[AU]UAAA-3' motif in vitro. Binds optimally to RNA containing 5'-[AU]UAA-3' as a bipartite motif spaced by more than 15 nucleotides. Binds poly(A). RNA-binding abilities are down-regulated by tyrosine kinase PTK6. Involved in splice site selection of vascular endothelial growth factor. In vitro regulates CD44 alternative splicing by direct binding to purine-rich exonic enhancer. Can regulate alternative splicing of neurexins NRXN1-3 in the laminin G-like domain 6 containing the evolutionary conserved neurexin alternative spliced segment 4 (AS4) involved in neurexin selective targeting to postsynaptic partners such as neuroligins and LRRTM family members. Targeted, cell-type specific splicing regulation of NRXN1 at AS4 is involved in neuronal glutamatergic synapse function and plasticity. May regulate expression of KHDRBS2/SLIM-1 in defined brain neuron populations by modifying its alternative splicing. Can bind FABP9 mRNA. May play a role as a negative regulator of cell growth. Inhibits cell proliferation.; (Microbial infection) Involved in post-transcriptional regulation of HIV-1 gene expression.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
HHS-475 Probe Info |
 |
Y5(0.85) | LDD0264 | [1] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [2] | |
|
HHS-465 Probe Info |
 |
N.A. | LDD2240 | [3] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C139 Probe Info |
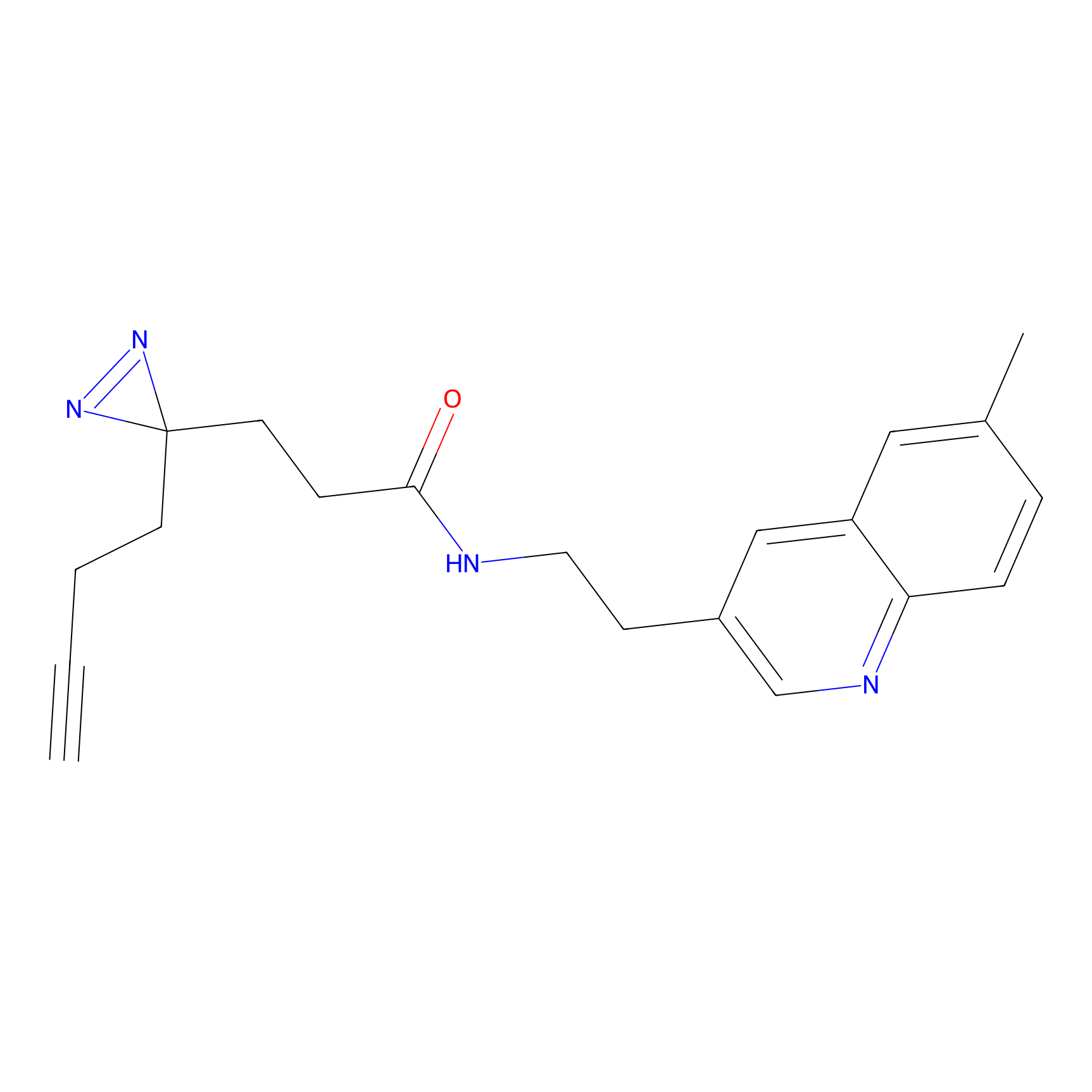 |
11.39 | LDD1821 | [4] | |
|
C165 Probe Info |
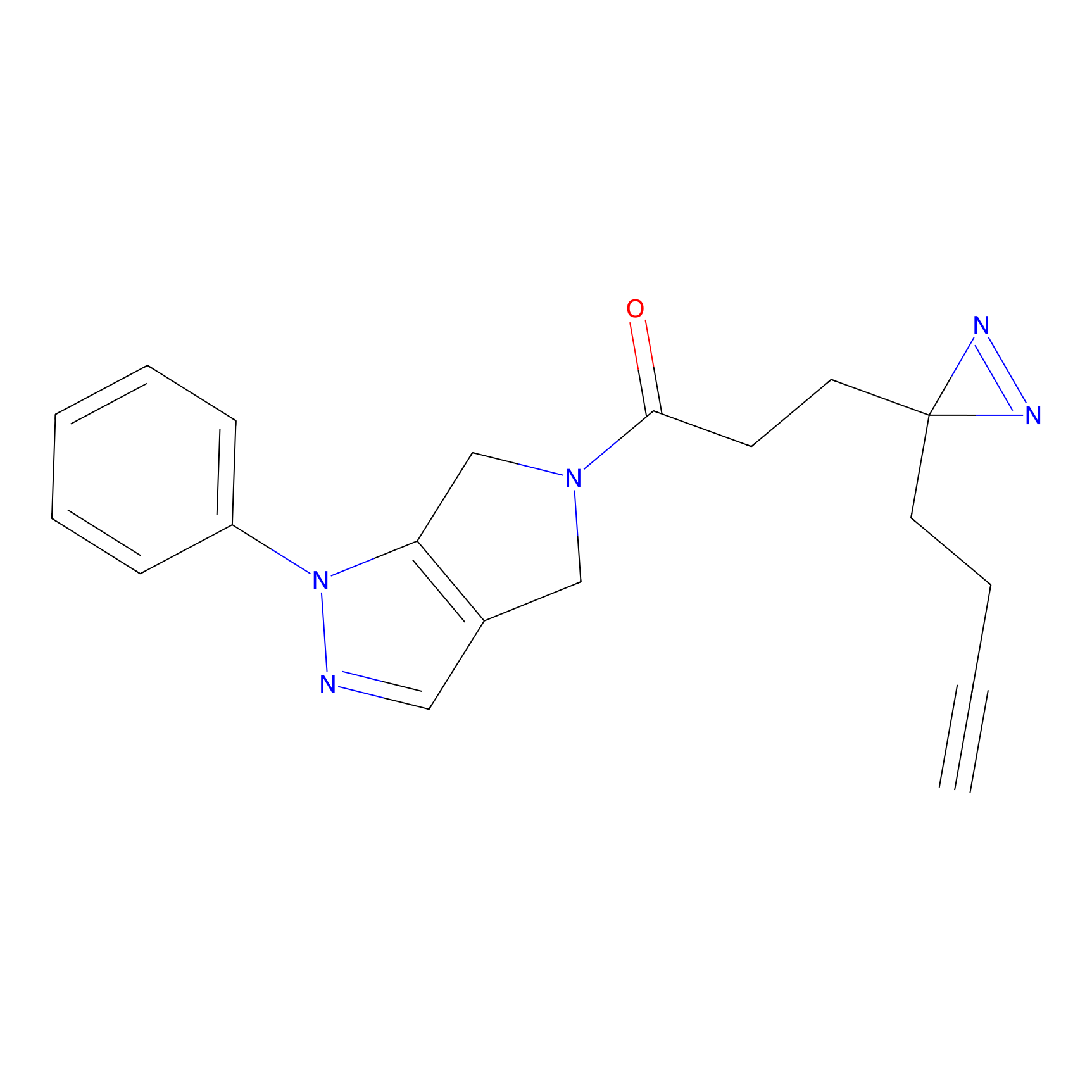 |
16.22 | LDD1845 | [4] | |
|
C293 Probe Info |
 |
17.15 | LDD1963 | [4] | |
|
C296 Probe Info |
 |
11.88 | LDD1966 | [4] | |
|
C313 Probe Info |
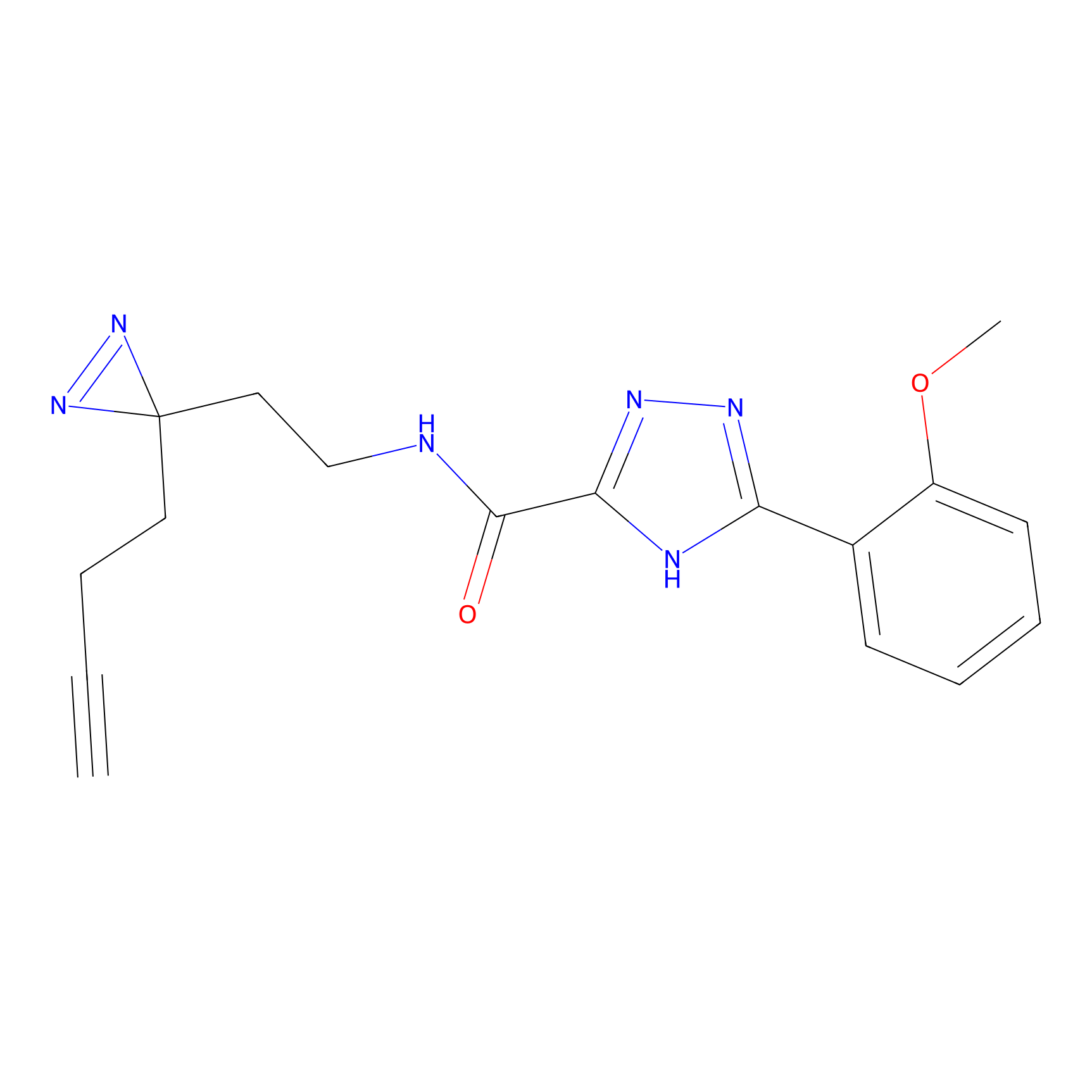 |
14.42 | LDD1980 | [4] | |
|
C314 Probe Info |
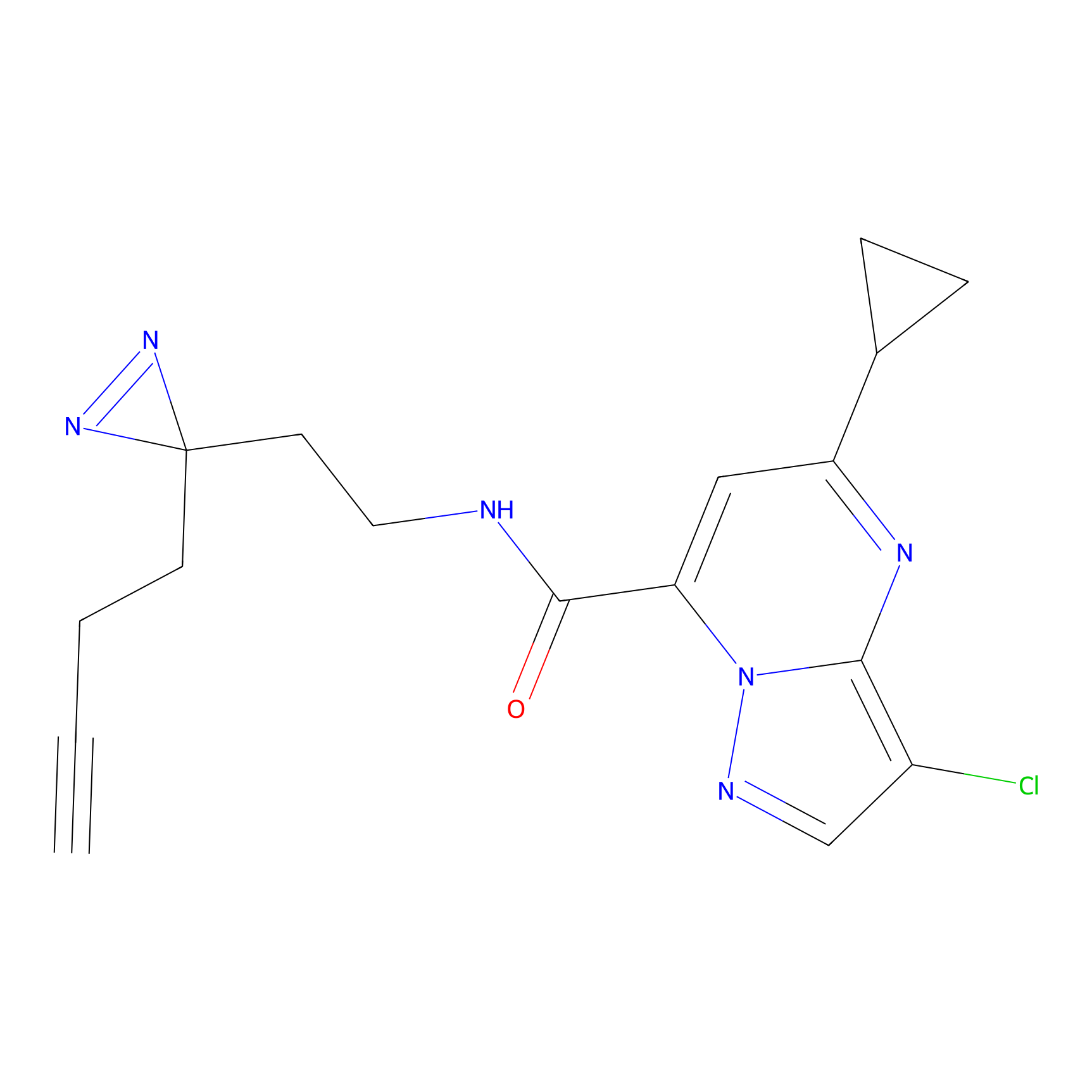 |
12.38 | LDD1981 | [4] | |
|
C346 Probe Info |
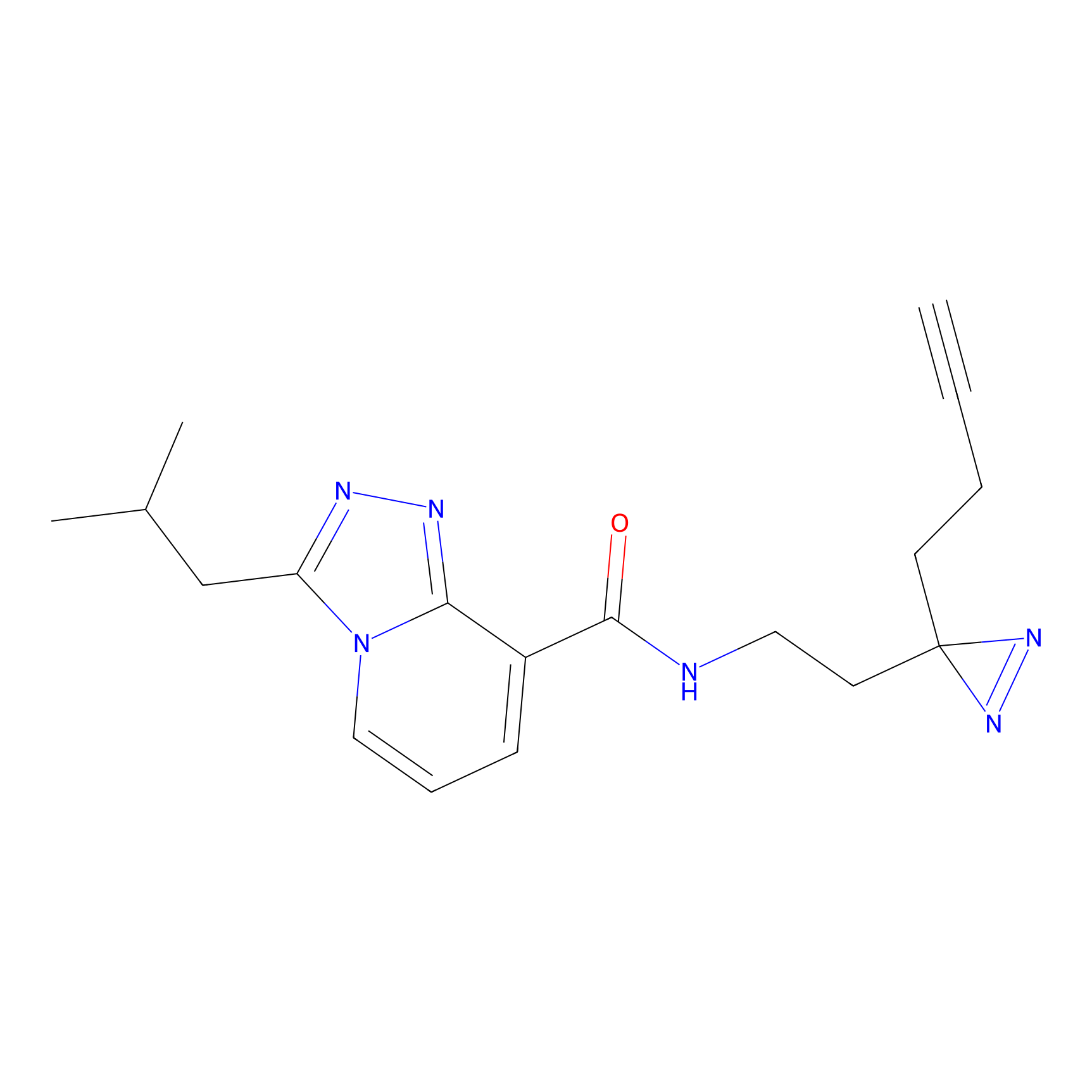 |
11.39 | LDD2007 | [4] | |
|
C355 Probe Info |
 |
23.43 | LDD2016 | [4] | |
|
C383 Probe Info |
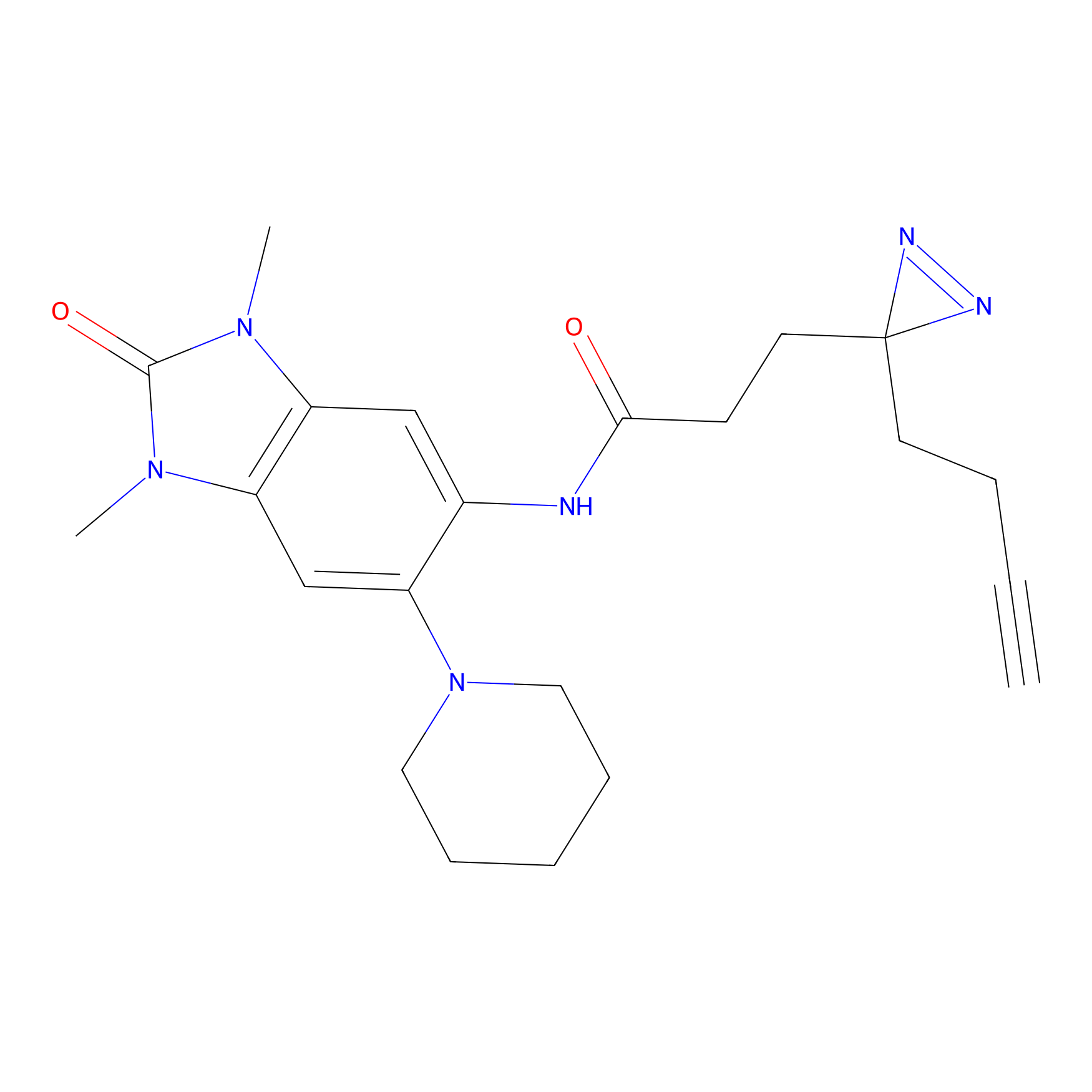 |
13.93 | LDD2042 | [4] | |
Competitor(s) Related to This Target
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
Transcription factor
Other
References
