Details of the Target
General Information of Target
| Target ID | LDTP00383 | |||||
|---|---|---|---|---|---|---|
| Target Name | AP-3 complex subunit delta-1 (AP3D1) | |||||
| Gene Name | AP3D1 | |||||
| Gene ID | 8943 | |||||
| Synonyms |
AP-3 complex subunit delta-1; AP-3 complex subunit delta; Adaptor-related protein complex 3 subunit delta-1; Delta-adaptin |
|||||
| 3D Structure | ||||||
| Sequence |
MALKMVKGSIDRMFDKNLQDLVRGIRNHKEDEAKYISQCIDEIKQELKQDNIAVKANAVC
KLTYLQMLGYDISWAAFNIIEVMSASKFTFKRIGYLAASQSFHEGTDVIMLTTNQIRKDL SSPSQYDTGVALTGLSCFVTPDLARDLANDIMTLMSHTKPYIRKKAVLIMYKVFLKYPES LRPAFPRLKEKLEDPDPGVQSAAVNVICELARRNPKNYLSLAPLFFKLMTSSTNNWVLIK IIKLFGALTPLEPRLGKKLIEPLTNLIHSTSAMSLLYECVNTVIAVLISLSSGMPNHSAS IQLCVQKLRILIEDSDQNLKYLGLLAMSKILKTHPKSVQSHKDLILQCLDDKDESIRLRA LDLLYGMVSKKNLMEIVKKLMTHVDKAEGTTYRDELLTKIIDICSQSNYQYITNFEWYIS ILVELTRLEGTRHGHLIAAQMLDVAIRVKAIRKFAVSQMSALLDSAHLLASSTQRNGICE VLYAAAWICGEFSEHLQEPHHTLEAMLRPRVTTLPGHIQAVYVQNVVKLYASILQQKEQA GEAEGAQAVTQLMVDRLPQFVQSADLEVQERASCILQLVKHIQKLQAKDVPVAEEVSALF AGELNPVAPKAQKKVPVPEGLDLDAWINEPLSDSESEDERPRAVFHEEEQRRPKHRPSEA DEEELARRREARKQEQANNPFYIKSSPSPQKRYQDTPGVEHIPVVQIDLSVPLKVPGLPM SDQYVKLEEERRHRQKLEKDKRRKKRKEKEKKGKRRHSSLPTESDEDIAPAQQVDIVTEE MPENALPSDEDDKDPNDPYRALDIDLDKPLADSEKLPIQKHRNTETSKSPEKDVPMVEKK SKKPKKKEKKHKEKERDKEKKKEKEKKKSPKPKKKKHRKEKEERTKGKKKSKKQPPGSEE AAGEPVQNGAPEEEQLPPESSYSLLAENSYVKMTCDIRGSLQEDSQVTVAIVLENRSSSI LKGMELSVLDSLNARMARPQGSSVHDGVPVPFQLPPGVSNEAQYVFTIQSIVMAQKLKGT LSFIAKNDEGATHEKLDFRLHFSCSSYLITTPCYSDAFAKLLESGDLSMSSIKVDGIRMS FQNLLAKICFHHHFSVVERVDSCASMYSRSIQGHHVCLLVKKGENSVSVDGKCSDSTLLS NLLEEMKATLAKC |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Adaptor complexes large subunit family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Part of the AP-3 complex, an adaptor-related complex which is not clathrin-associated. The complex is associated with the Golgi region as well as more peripheral structures. It facilitates the budding of vesicles from the Golgi membrane and may be directly involved in trafficking to lysosomes. Involved in process of CD8+ T-cell and NK cell degranulation. In concert with the BLOC-1 complex, AP-3 is required to target cargos into vesicles assembled at cell bodies for delivery into neurites and nerve terminals.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
10.88 | LDD0402 | [1] | |
|
A-EBA Probe Info |
 |
3.15 | LDD0215 | [2] | |
|
STPyne Probe Info |
 |
K580(8.71); K584(9.70); K7(8.25); K820(1.15) | LDD0277 | [3] | |
|
DBIA Probe Info |
 |
C60(4.35); C574(7.55); C997(1.91); C971(2.09) | LDD3311 | [4] | |
|
BTD Probe Info |
 |
C60(1.52); C1153(1.38) | LDD1700 | [5] | |
|
AHL-Pu-1 Probe Info |
 |
C574(2.53) | LDD0168 | [6] | |
|
HHS-465 Probe Info |
 |
Y682(1.88) | LDD2237 | [7] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [8] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C574(0.00); C348(0.00); C208(0.00); C1117(0.00) | LDD0038 | [9] | |
|
IA-alkyne Probe Info |
 |
C574(0.00); C348(0.00); C1133(0.00) | LDD0032 | [10] | |
|
IPIAA_L Probe Info |
 |
C1133(0.00); C39(0.00) | LDD0031 | [11] | |
|
Lodoacetamide azide Probe Info |
 |
C574(0.00); C1133(0.00); C348(0.00); C208(0.00) | LDD0037 | [9] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [12] | |
|
TFBX Probe Info |
 |
C1103(0.00); C574(0.00) | LDD0027 | [13] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [14] | |
|
Compound 10 Probe Info |
 |
C1103(0.00); C574(0.00) | LDD2216 | [15] | |
|
IPM Probe Info |
 |
C1103(0.00); C574(0.00) | LDD0005 | [14] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [14] | |
|
1c-yne Probe Info |
 |
K588(0.00); K610(0.00) | LDD0228 | [16] | |
|
Acrolein Probe Info |
 |
C348(0.00); C208(0.00); C574(0.00); C935(0.00) | LDD0217 | [17] | |
|
Methacrolein Probe Info |
 |
C574(0.00); C348(0.00) | LDD0218 | [17] | |
|
W1 Probe Info |
 |
C39(0.00); C1103(0.00); C348(0.00) | LDD0236 | [18] | |
|
AOyne Probe Info |
 |
9.30 | LDD0443 | [19] | |
|
NAIA_5 Probe Info |
 |
C574(0.00); C1133(0.00); C1089(0.00); C208(0.00) | LDD2223 | [12] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C095 Probe Info |
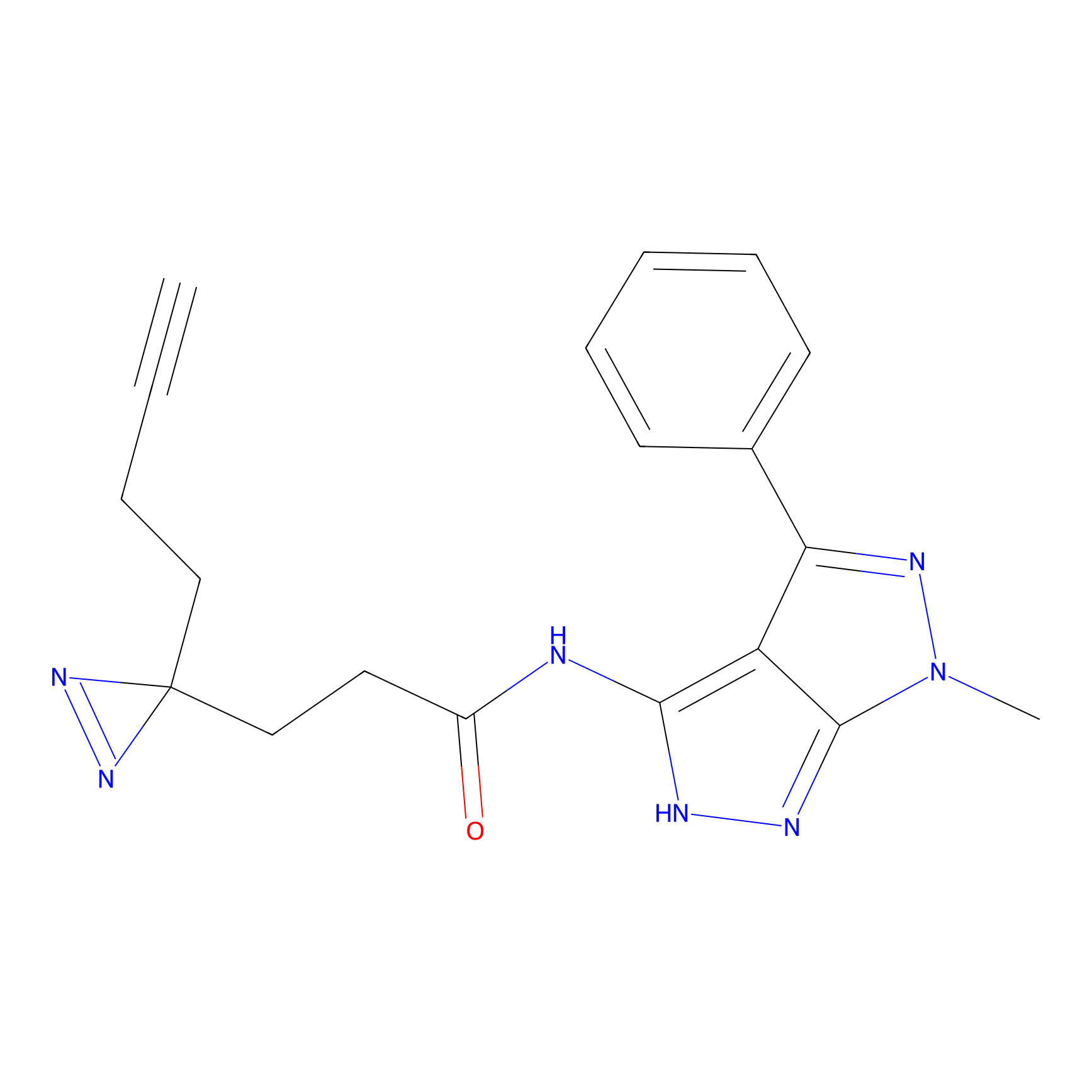 |
5.54 | LDD1786 | [20] | |
|
C183 Probe Info |
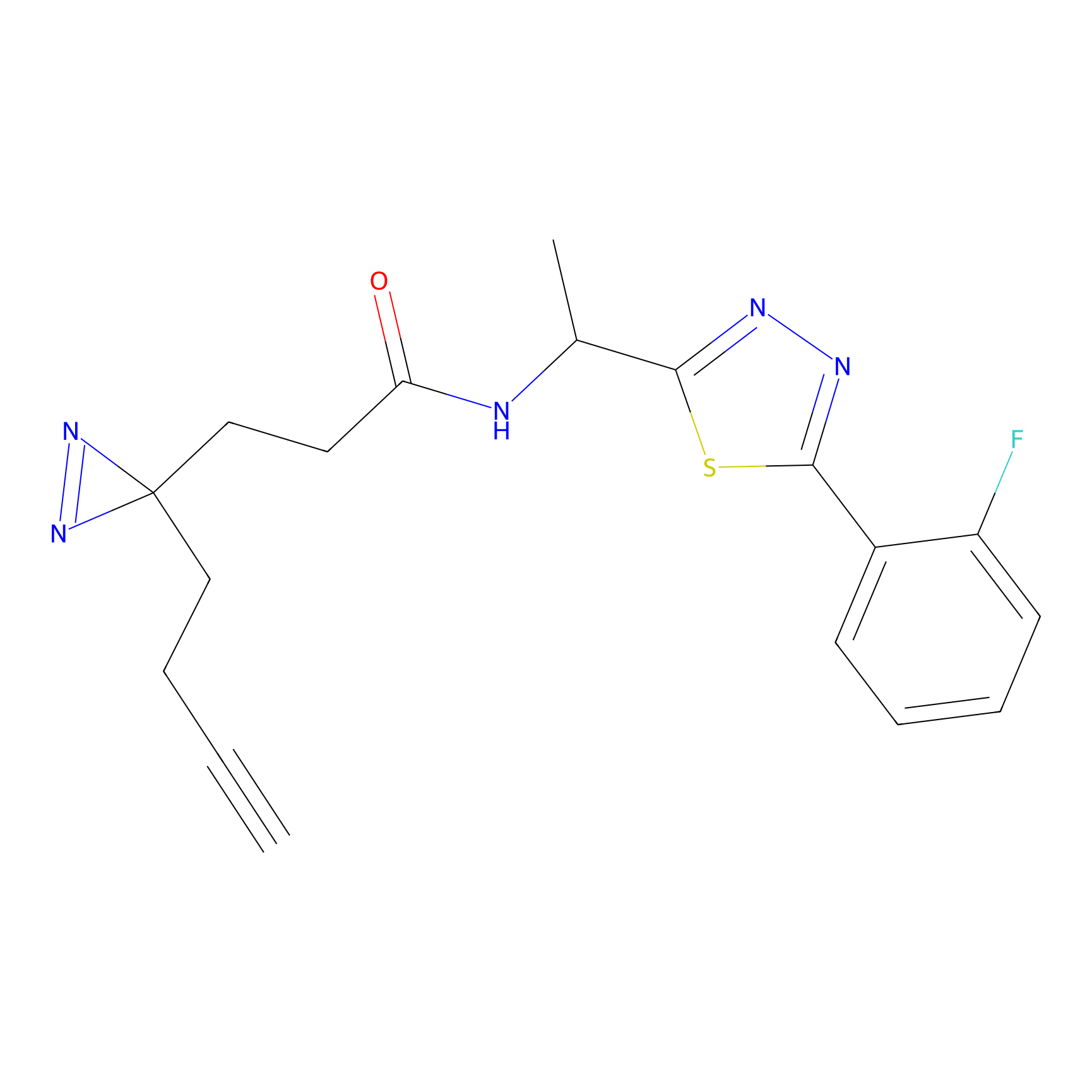 |
9.58 | LDD1861 | [20] | |
|
C196 Probe Info |
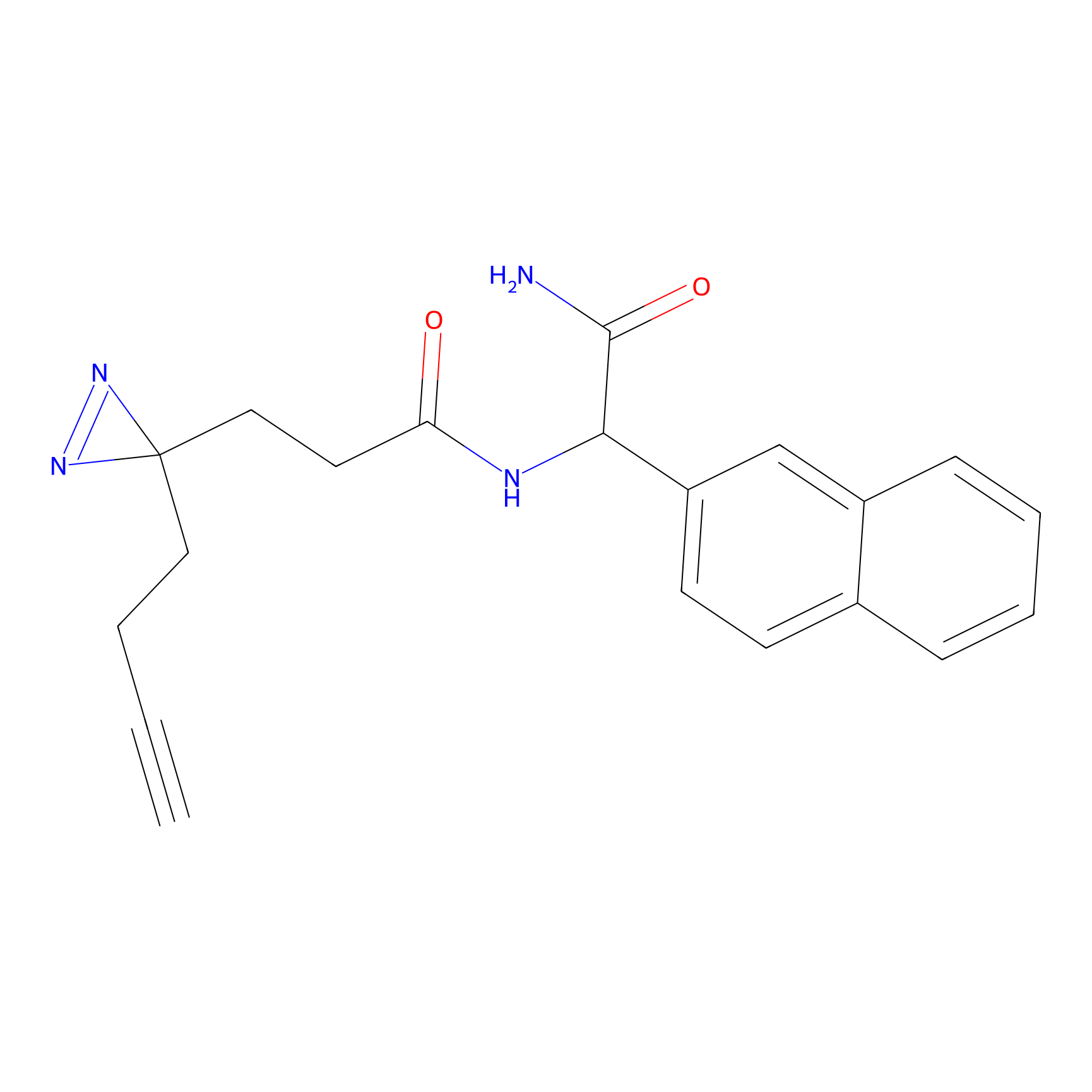 |
13.55 | LDD1872 | [20] | |
|
C243 Probe Info |
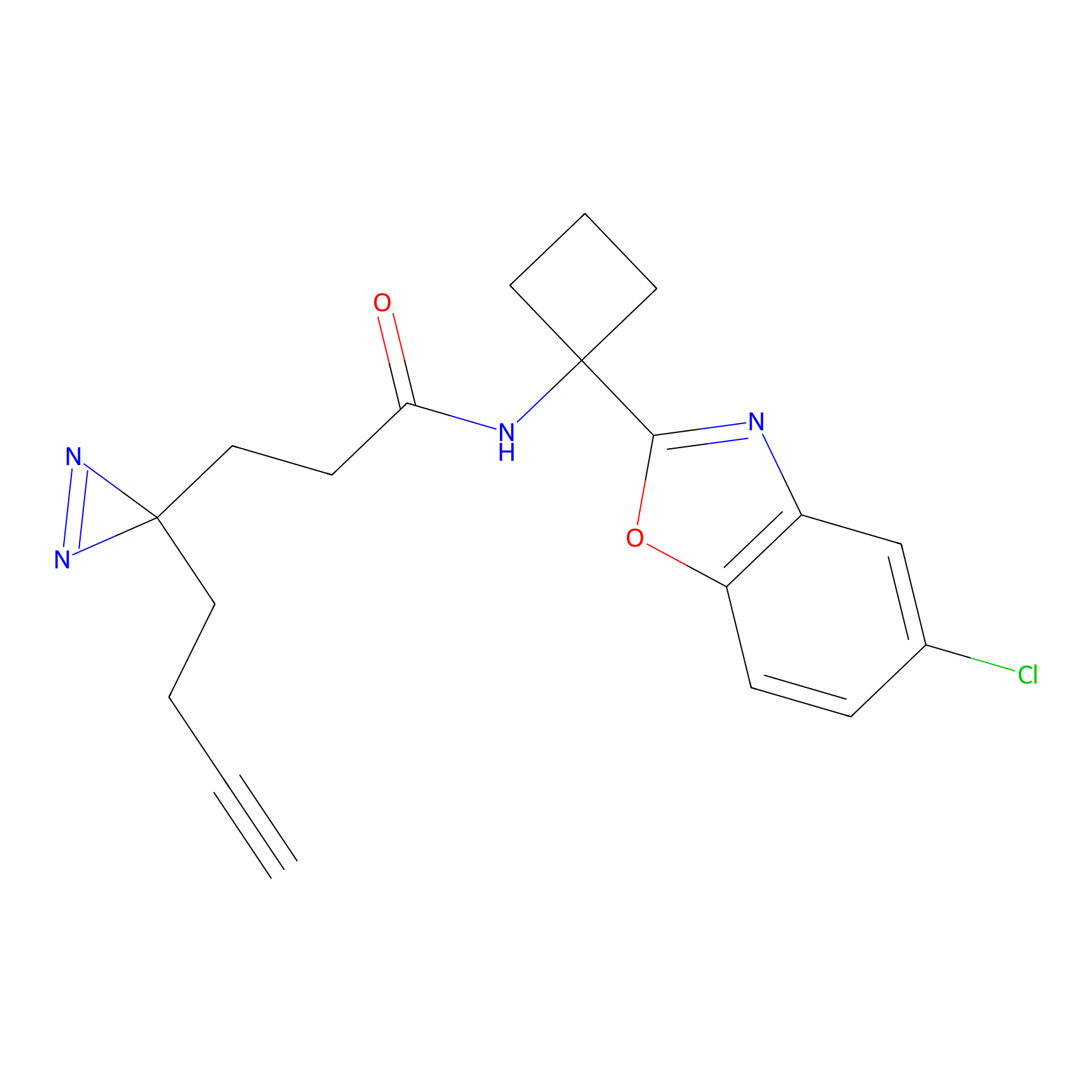 |
12.21 | LDD1916 | [20] | |
|
C349 Probe Info |
 |
8.94 | LDD2010 | [20] | |
|
FFF probe4 Probe Info |
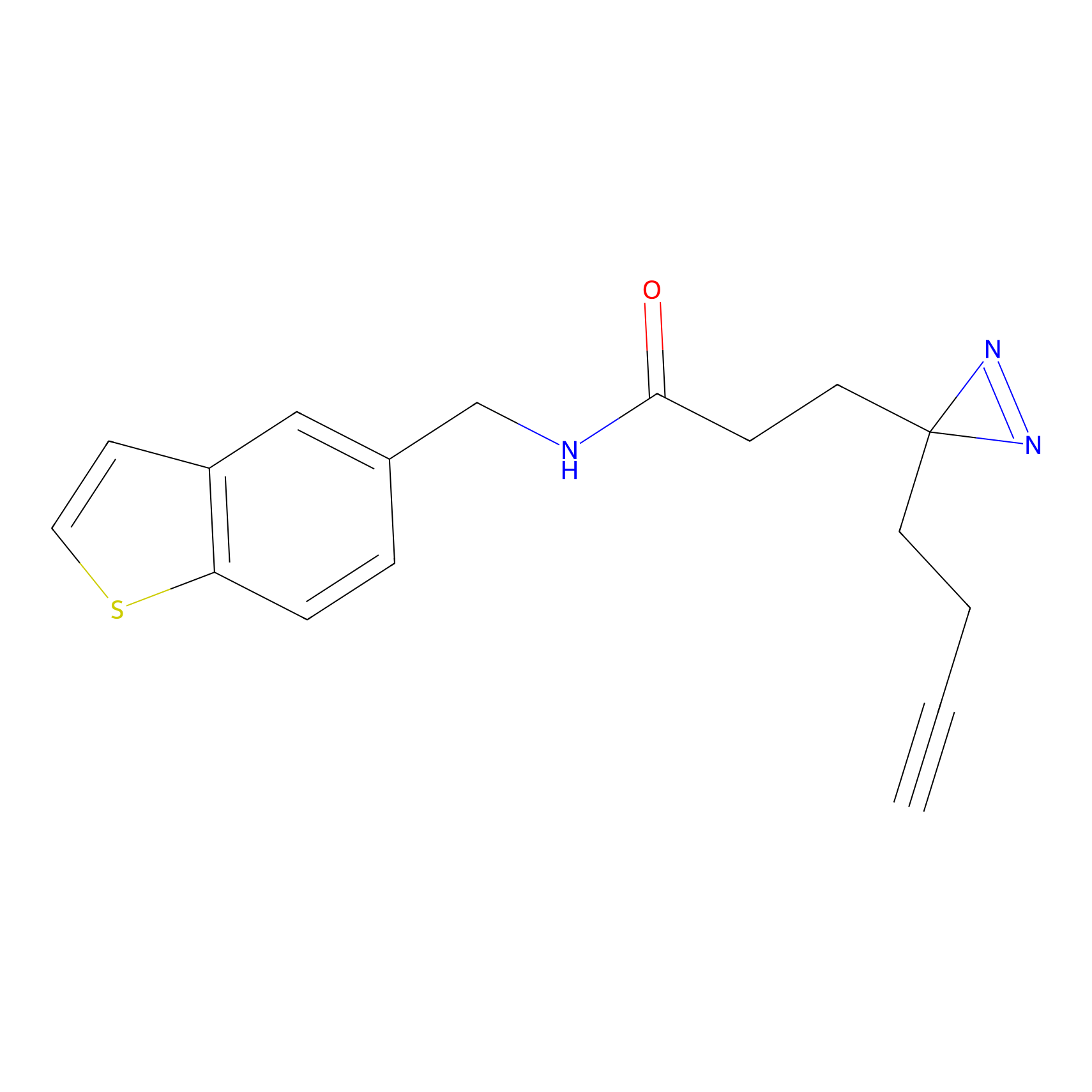 |
12.65 | LDD0466 | [21] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [22] | |
|
OEA-DA Probe Info |
 |
3.07 | LDD0046 | [23] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C574(0.36) | LDD2142 | [5] |
| LDCM0502 | 1-(Cyanoacetyl)piperidine | MDA-MB-231 | C60(0.63) | LDD2095 | [5] |
| LDCM0537 | 2-Cyano-N,N-dimethylacetamide | MDA-MB-231 | C60(0.73) | LDD2130 | [5] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C60(0.99); C1153(1.06) | LDD2117 | [5] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C60(1.12); C1153(0.81) | LDD2152 | [5] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C60(1.09) | LDD2132 | [5] |
| LDCM0025 | 4SU-RNA | HEK-293T | C574(2.53) | LDD0168 | [6] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C574(8.42); C1103(2.51) | LDD0169 | [6] |
| LDCM0545 | Acetamide | MDA-MB-231 | C574(0.49) | LDD2138 | [5] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C60(1.15) | LDD2113 | [5] |
| LDCM0156 | Aniline | NCI-H1299 | 12.12 | LDD0403 | [1] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C574(0.53) | LDD2091 | [5] |
| LDCM0108 | Chloroacetamide | HeLa | C574(0.00); C348(0.00); H655(0.00) | LDD0222 | [17] |
| LDCM0632 | CL-Sc | Hep-G2 | C574(1.23); C1103(0.59) | LDD2227 | [12] |
| LDCM0634 | CY-0357 | Hep-G2 | C1103(0.52) | LDD2228 | [12] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C574(7.17); C60(0.89) | LDD1702 | [5] |
| LDCM0625 | F8 | Ramos | C1103(1.03); C574(0.65) | LDD2187 | [24] |
| LDCM0572 | Fragment10 | Ramos | C1103(1.47); C574(0.86) | LDD2189 | [24] |
| LDCM0573 | Fragment11 | Ramos | C1103(0.79); C574(20.00) | LDD2190 | [24] |
| LDCM0574 | Fragment12 | Ramos | C1103(0.95); C574(0.83) | LDD2191 | [24] |
| LDCM0575 | Fragment13 | Ramos | C1103(1.05); C574(0.93) | LDD2192 | [24] |
| LDCM0576 | Fragment14 | Ramos | C1103(1.00); C574(1.94) | LDD2193 | [24] |
| LDCM0579 | Fragment20 | Ramos | C1103(0.84); C574(0.64) | LDD2194 | [24] |
| LDCM0580 | Fragment21 | Ramos | C1103(1.10); C574(0.99) | LDD2195 | [24] |
| LDCM0582 | Fragment23 | Ramos | C1103(0.91); C574(0.39) | LDD2196 | [24] |
| LDCM0578 | Fragment27 | Ramos | C1103(1.02); C574(0.97) | LDD2197 | [24] |
| LDCM0586 | Fragment28 | Ramos | C1103(0.97); C574(1.88) | LDD2198 | [24] |
| LDCM0588 | Fragment30 | Ramos | C1103(1.79); C574(1.06) | LDD2199 | [24] |
| LDCM0589 | Fragment31 | Ramos | C1103(1.19) | LDD2200 | [24] |
| LDCM0590 | Fragment32 | Ramos | C1103(1.08); C574(0.78) | LDD2201 | [24] |
| LDCM0468 | Fragment33 | Ramos | C1103(1.08); C574(1.05) | LDD2202 | [24] |
| LDCM0596 | Fragment38 | Ramos | C1103(0.93); C574(0.88) | LDD2203 | [24] |
| LDCM0566 | Fragment4 | Ramos | C1103(1.14); C574(0.87) | LDD2184 | [24] |
| LDCM0610 | Fragment52 | Ramos | C1103(1.34); C574(2.12) | LDD2204 | [24] |
| LDCM0614 | Fragment56 | Ramos | C1103(1.52); C574(1.47) | LDD2205 | [24] |
| LDCM0569 | Fragment7 | Ramos | C1103(0.92); C574(0.71) | LDD2186 | [24] |
| LDCM0571 | Fragment9 | Ramos | C1103(1.14); C574(0.81) | LDD2188 | [24] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [17] |
| LDCM0022 | KB02 | Ramos | C1103(1.19); C574(0.72) | LDD2182 | [24] |
| LDCM0023 | KB03 | MDA-MB-231 | C574(19.65); C60(0.81) | LDD1701 | [5] |
| LDCM0024 | KB05 | G361 | C60(4.35); C574(7.55); C997(1.91); C971(2.09) | LDD3311 | [4] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C60(0.85); C574(0.86) | LDD2121 | [5] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [17] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C60(0.52); C574(0.81) | LDD2089 | [5] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C60(0.92); C1153(1.01); C39(1.45) | LDD2093 | [5] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C60(1.01) | LDD2097 | [5] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C60(1.00); C1153(1.02) | LDD2099 | [5] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C60(1.14) | LDD2101 | [5] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C60(1.01); C1153(0.99) | LDD2107 | [5] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C1153(0.53); C935(1.33) | LDD2109 | [5] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C60(1.07) | LDD2111 | [5] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C574(0.89) | LDD2114 | [5] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C60(2.64) | LDD2119 | [5] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C60(0.94); C1153(0.89) | LDD2120 | [5] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C60(0.84); C1153(0.75) | LDD2123 | [5] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C60(0.98); C1103(0.88) | LDD2125 | [5] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C60(1.01); C1153(1.12) | LDD2127 | [5] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C60(2.40); C1153(0.88) | LDD2128 | [5] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C60(1.05); C1153(0.88) | LDD2129 | [5] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C60(0.59) | LDD2134 | [5] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C1153(1.26); C39(2.18) | LDD2135 | [5] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C60(1.06) | LDD2136 | [5] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C60(1.06); C1153(0.88) | LDD2137 | [5] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C60(1.52); C1153(1.38) | LDD1700 | [5] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C60(0.65); C1153(0.47) | LDD2140 | [5] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C574(0.63) | LDD2141 | [5] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C60(1.26); C1153(0.90) | LDD2143 | [5] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C60(2.74) | LDD2144 | [5] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C60(0.89); C1153(0.99) | LDD2146 | [5] |
| LDCM0553 | Nucleophilic fragment 6b | MDA-MB-231 | C1153(2.78) | LDD2147 | [5] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C60(0.64) | LDD2148 | [5] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C574(0.07) | LDD2206 | [25] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C574(1.13) | LDD2207 | [25] |
References
