Details of the Target
General Information of Target
| Target ID | LDTP09861 | |||||
|---|---|---|---|---|---|---|
| Target Name | General transcription factor IIH subunit 4 (GTF2H4) | |||||
| Gene Name | GTF2H4 | |||||
| Gene ID | 2968 | |||||
| Synonyms |
General transcription factor IIH subunit 4; Basic transcription factor 2 52 kDa subunit; BTF2 p52; General transcription factor IIH polypeptide 4; TFIIH basal transcription factor complex p52 subunit |
|||||
| 3D Structure | ||||||
| Sequence |
MLWKITDNVKYEEDCEDRHDGSSNGNPRVPHLSSAGQHLYSPAPPLSHTGVAEYQPPPYF
PPPYQQLAYSQSADPYSHLGEAYAAAINPLHQPAPTGSQQQAWPGRQSQEGAGLPSHHGR PAGLLPHLSGLEAGAVSARRDAYRRSDLLLPHAHALDAAGLAENLGLHDMPHQMDEVQNV DDQHLLLHDQTVIRKGPISMTKNPLNLPCQKELVGAVMNPTEVFCSVPGRLSLLSSTSKY KVTVAEVQRRLSPPECLNASLLGGVLRRAKSKNGGRSLREKLDKIGLNLPAGRRKAAHVT LLTSLVEGEAVHLARDFAYVCEAEFPSKPVAEYLTRPHLGGRNEMAARKNMLLAAQQLCK EFTELLSQDRTPHGTSRLAPVLETNIQNCLSHFSLITHGFGSQAICAAVSALQNYIKEAL IVIDKSYMNPGDQSPADSNKTLEKMEKHRK |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
TFB2 family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Component of the general transcription and DNA repair factor IIH (TFIIH) core complex, which is involved in general and transcription-coupled nucleotide excision repair (NER) of damaged DNA and, when complexed to CAK, in RNA transcription by RNA polymerase II. In NER, TFIIH acts by opening DNA around the lesion to allow the excision of the damaged oligonucleotide and its replacement by a new DNA fragment. In transcription, TFIIH has an essential role in transcription initiation. When the pre-initiation complex (PIC) has been established, TFIIH is required for promoter opening and promoter escape. Phosphorylation of the C-terminal tail (CTD) of the largest subunit of RNA polymerase II by the kinase module CAK controls the initiation of transcription.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| COLO792 | SNV: p.S147F | DBIA Probe Info | |||
| LNCaP clone FGC | SNV: p.R274K | DBIA Probe Info | |||
| MCAS | SNV: p.R46T | DBIA Probe Info | |||
| MOLT4 | SNV: p.M57I | IA-alkyne Probe Info | |||
| OVCAR8 | SNV: p.L327F | . | |||
| SKES1 | SNV: p.H366Q | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
BTD Probe Info |
 |
C16(0.68) | LDD2089 | [2] | |
|
Curcusone 37 Probe Info |
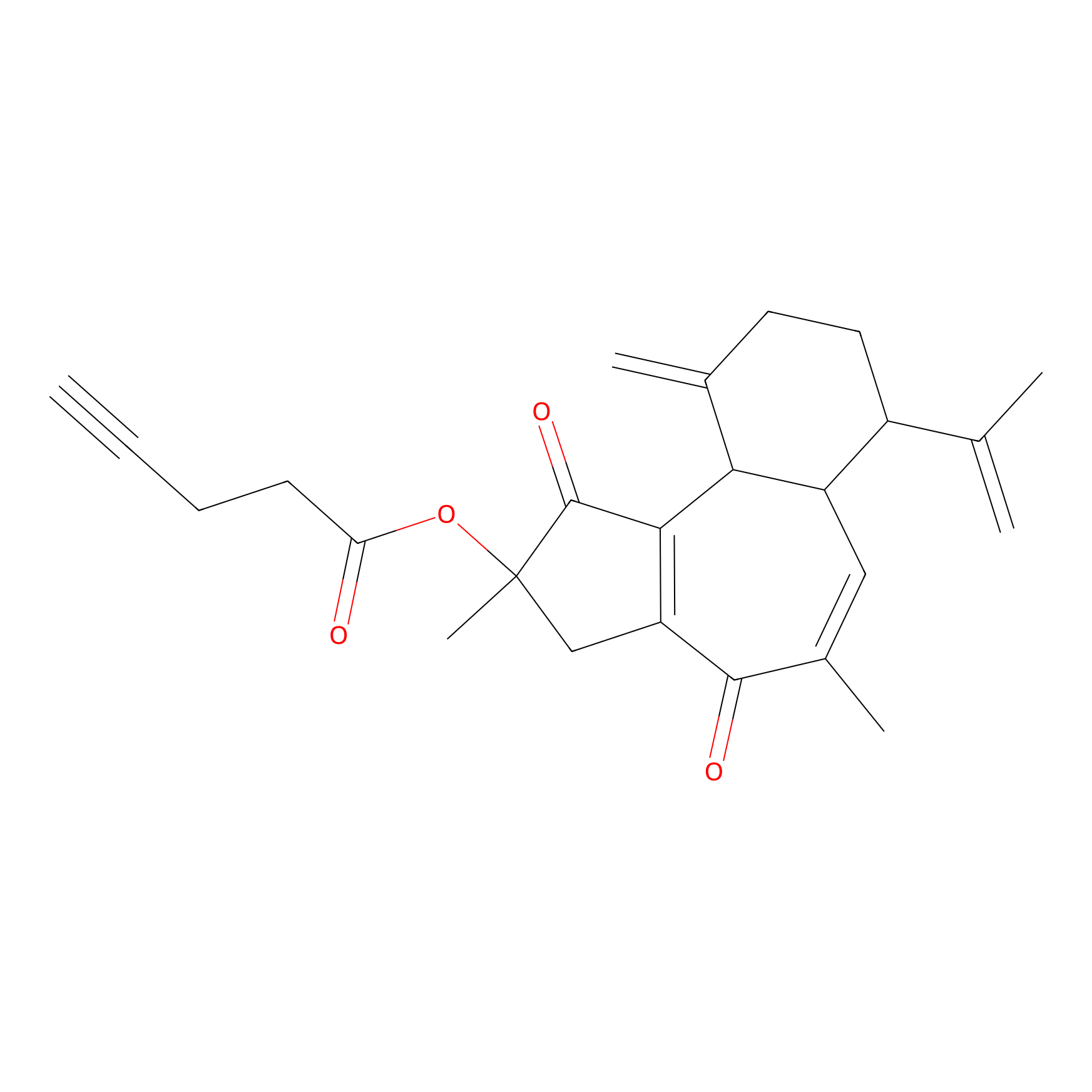 |
2.54 | LDD0188 | [3] | |
|
AHL-Pu-1 Probe Info |
 |
C16(2.13) | LDD0168 | [4] | |
|
DBIA Probe Info |
 |
C16(4.74) | LDD0209 | [5] | |
|
Acrolein Probe Info |
 |
C41(0.00); C16(0.00) | LDD0222 | [6] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [7] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [8] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [8] | |
|
IPM Probe Info |
 |
N.A. | LDD0147 | [9] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [9] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [10] | |
|
NAIA_5 Probe Info |
 |
C16(0.00); C41(0.00) | LDD2223 | [11] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C160 Probe Info |
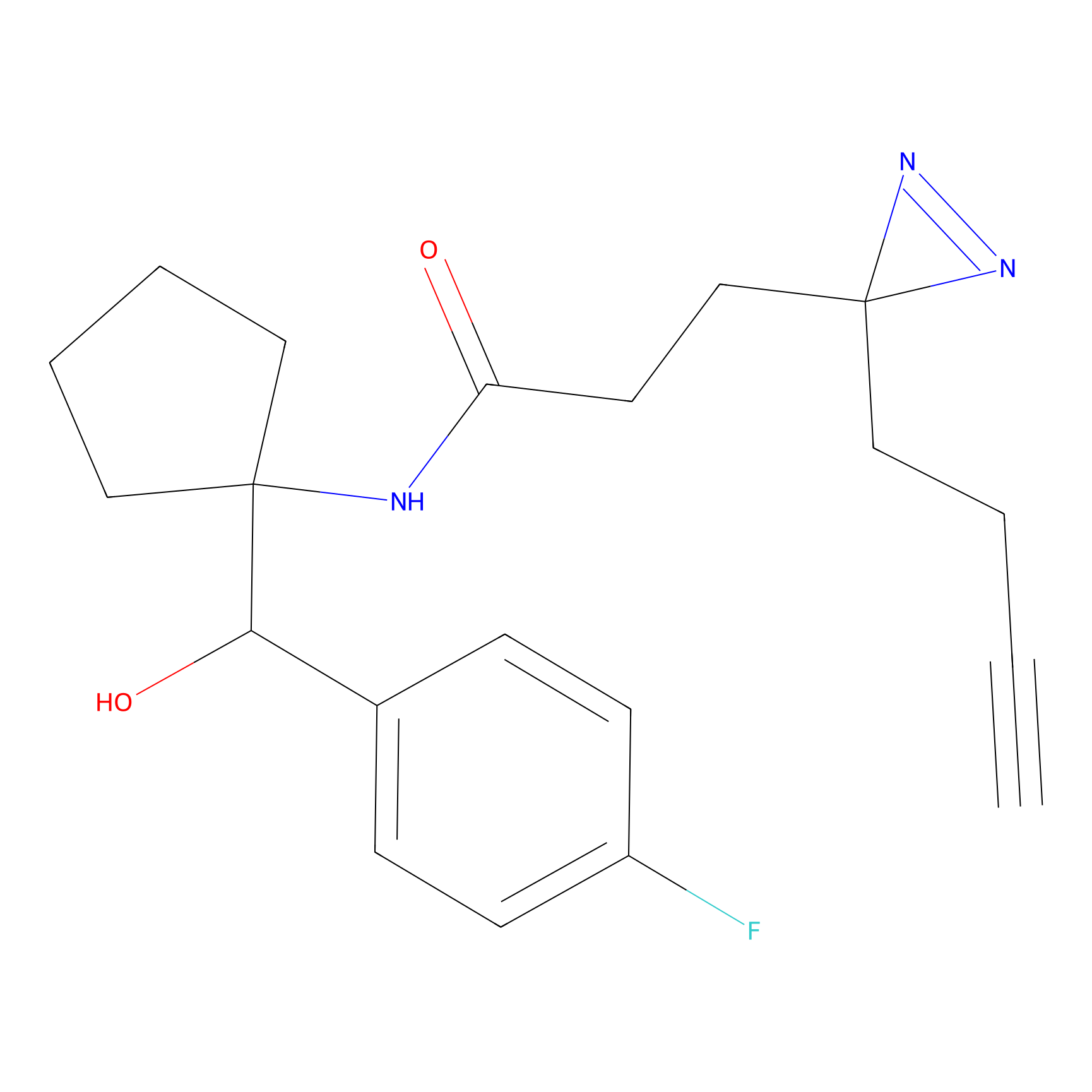 |
7.21 | LDD1840 | [12] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C16(0.97) | LDD2117 | [2] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C16(1.18) | LDD2152 | [2] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C16(0.52) | LDD2132 | [2] |
| LDCM0025 | 4SU-RNA | HEK-293T | C16(2.13) | LDD0168 | [4] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C16(2.45) | LDD0169 | [4] |
| LDCM0214 | AC1 | HEK-293T | C41(1.02) | LDD1507 | [13] |
| LDCM0215 | AC10 | HEK-293T | C41(0.94) | LDD1508 | [13] |
| LDCM0226 | AC11 | HEK-293T | C41(0.97) | LDD1509 | [13] |
| LDCM0237 | AC12 | HEK-293T | C41(0.98) | LDD1510 | [13] |
| LDCM0259 | AC14 | HEK-293T | C41(0.93) | LDD1512 | [13] |
| LDCM0270 | AC15 | HEK-293T | C41(1.00) | LDD1513 | [13] |
| LDCM0276 | AC17 | HEK-293T | C41(0.96) | LDD1515 | [13] |
| LDCM0277 | AC18 | HEK-293T | C41(0.94) | LDD1516 | [13] |
| LDCM0278 | AC19 | HEK-293T | C41(0.91) | LDD1517 | [13] |
| LDCM0279 | AC2 | HEK-293T | C41(0.99) | LDD1518 | [13] |
| LDCM0280 | AC20 | HEK-293T | C41(0.97) | LDD1519 | [13] |
| LDCM0281 | AC21 | HEK-293T | C41(0.84) | LDD1520 | [13] |
| LDCM0282 | AC22 | HEK-293T | C41(0.96) | LDD1521 | [13] |
| LDCM0283 | AC23 | HEK-293T | C41(0.93) | LDD1522 | [13] |
| LDCM0284 | AC24 | HEK-293T | C41(1.14) | LDD1523 | [13] |
| LDCM0285 | AC25 | HEK-293T | C41(0.98) | LDD1524 | [13] |
| LDCM0286 | AC26 | HEK-293T | C41(0.92) | LDD1525 | [13] |
| LDCM0287 | AC27 | HEK-293T | C41(0.92) | LDD1526 | [13] |
| LDCM0288 | AC28 | HEK-293T | C41(0.92) | LDD1527 | [13] |
| LDCM0289 | AC29 | HEK-293T | C41(0.79) | LDD1528 | [13] |
| LDCM0290 | AC3 | HEK-293T | C41(0.97) | LDD1529 | [13] |
| LDCM0291 | AC30 | HEK-293T | C41(0.79) | LDD1530 | [13] |
| LDCM0292 | AC31 | HEK-293T | C41(0.96) | LDD1531 | [13] |
| LDCM0293 | AC32 | HEK-293T | C41(0.86) | LDD1532 | [13] |
| LDCM0294 | AC33 | HEK-293T | C41(0.93) | LDD1533 | [13] |
| LDCM0295 | AC34 | HEK-293T | C41(0.95) | LDD1534 | [13] |
| LDCM0296 | AC35 | HEK-293T | C41(0.99) | LDD1535 | [13] |
| LDCM0297 | AC36 | HEK-293T | C41(0.88) | LDD1536 | [13] |
| LDCM0298 | AC37 | HEK-293T | C41(0.99) | LDD1537 | [13] |
| LDCM0299 | AC38 | HEK-293T | C41(0.92) | LDD1538 | [13] |
| LDCM0300 | AC39 | HEK-293T | C41(0.95) | LDD1539 | [13] |
| LDCM0301 | AC4 | HEK-293T | C41(0.93) | LDD1540 | [13] |
| LDCM0302 | AC40 | HEK-293T | C41(0.85) | LDD1541 | [13] |
| LDCM0303 | AC41 | HEK-293T | C41(0.97) | LDD1542 | [13] |
| LDCM0304 | AC42 | HEK-293T | C41(0.93) | LDD1543 | [13] |
| LDCM0305 | AC43 | HEK-293T | C41(0.99) | LDD1544 | [13] |
| LDCM0306 | AC44 | HEK-293T | C41(0.99) | LDD1545 | [13] |
| LDCM0307 | AC45 | HEK-293T | C41(0.88) | LDD1546 | [13] |
| LDCM0308 | AC46 | HEK-293T | C41(0.95) | LDD1547 | [13] |
| LDCM0309 | AC47 | HEK-293T | C41(0.92) | LDD1548 | [13] |
| LDCM0310 | AC48 | HEK-293T | C41(1.17) | LDD1549 | [13] |
| LDCM0311 | AC49 | HEK-293T | C41(1.01) | LDD1550 | [13] |
| LDCM0312 | AC5 | HEK-293T | C41(0.98) | LDD1551 | [13] |
| LDCM0313 | AC50 | HEK-293T | C41(0.99) | LDD1552 | [13] |
| LDCM0314 | AC51 | HEK-293T | C41(0.97) | LDD1553 | [13] |
| LDCM0315 | AC52 | HEK-293T | C41(0.99) | LDD1554 | [13] |
| LDCM0316 | AC53 | HEK-293T | C41(0.93) | LDD1555 | [13] |
| LDCM0317 | AC54 | HEK-293T | C41(0.94) | LDD1556 | [13] |
| LDCM0318 | AC55 | HEK-293T | C41(0.95) | LDD1557 | [13] |
| LDCM0319 | AC56 | HEK-293T | C41(1.12) | LDD1558 | [13] |
| LDCM0320 | AC57 | HEK-293T | C41(1.03) | LDD1559 | [13] |
| LDCM0321 | AC58 | HEK-293T | C41(1.02) | LDD1560 | [13] |
| LDCM0322 | AC59 | HEK-293T | C41(0.96) | LDD1561 | [13] |
| LDCM0323 | AC6 | HEK-293T | C41(0.98) | LDD1562 | [13] |
| LDCM0324 | AC60 | HEK-293T | C41(0.86) | LDD1563 | [13] |
| LDCM0325 | AC61 | HEK-293T | C41(0.94) | LDD1564 | [13] |
| LDCM0326 | AC62 | HEK-293T | C41(0.90) | LDD1565 | [13] |
| LDCM0327 | AC63 | HEK-293T | C41(1.05) | LDD1566 | [13] |
| LDCM0328 | AC64 | HEK-293T | C41(1.07) | LDD1567 | [13] |
| LDCM0334 | AC7 | HEK-293T | C41(1.10) | LDD1568 | [13] |
| LDCM0345 | AC8 | HEK-293T | C41(0.74) | LDD1569 | [13] |
| LDCM0248 | AKOS034007472 | HEK-293T | C41(0.96) | LDD1511 | [13] |
| LDCM0356 | AKOS034007680 | HEK-293T | C41(0.98) | LDD1570 | [13] |
| LDCM0275 | AKOS034007705 | HEK-293T | C41(0.87) | LDD1514 | [13] |
| LDCM0156 | Aniline | NCI-H1299 | 8.94 | LDD0403 | [1] |
| LDCM0108 | Chloroacetamide | HeLa | C41(0.00); C16(0.00) | LDD0222 | [6] |
| LDCM0367 | CL1 | HEK-293T | C41(1.02) | LDD1571 | [13] |
| LDCM0368 | CL10 | HEK-293T | C41(1.21) | LDD1572 | [13] |
| LDCM0369 | CL100 | HEK-293T | C41(0.95) | LDD1573 | [13] |
| LDCM0370 | CL101 | HEK-293T | C41(0.99) | LDD1574 | [13] |
| LDCM0371 | CL102 | HEK-293T | C41(1.11) | LDD1575 | [13] |
| LDCM0372 | CL103 | HEK-293T | C41(0.83) | LDD1576 | [13] |
| LDCM0373 | CL104 | HEK-293T | C41(0.89) | LDD1577 | [13] |
| LDCM0374 | CL105 | HEK-293T | C41(1.14) | LDD1578 | [13] |
| LDCM0375 | CL106 | HEK-293T | C41(0.99) | LDD1579 | [13] |
| LDCM0376 | CL107 | HEK-293T | C41(0.98) | LDD1580 | [13] |
| LDCM0377 | CL108 | HEK-293T | C41(0.98) | LDD1581 | [13] |
| LDCM0378 | CL109 | HEK-293T | C41(0.93) | LDD1582 | [13] |
| LDCM0379 | CL11 | HEK-293T | C41(1.15) | LDD1583 | [13] |
| LDCM0380 | CL110 | HEK-293T | C41(1.03) | LDD1584 | [13] |
| LDCM0381 | CL111 | HEK-293T | C41(0.88) | LDD1585 | [13] |
| LDCM0382 | CL112 | HEK-293T | C41(1.11) | LDD1586 | [13] |
| LDCM0383 | CL113 | HEK-293T | C41(1.10) | LDD1587 | [13] |
| LDCM0384 | CL114 | HEK-293T | C41(0.94) | LDD1588 | [13] |
| LDCM0385 | CL115 | HEK-293T | C41(0.91) | LDD1589 | [13] |
| LDCM0386 | CL116 | HEK-293T | C41(0.94) | LDD1590 | [13] |
| LDCM0387 | CL117 | HEK-293T | C41(1.02) | LDD1591 | [13] |
| LDCM0388 | CL118 | HEK-293T | C41(0.90) | LDD1592 | [13] |
| LDCM0389 | CL119 | HEK-293T | C41(1.03) | LDD1593 | [13] |
| LDCM0390 | CL12 | HEK-293T | C41(1.15) | LDD1594 | [13] |
| LDCM0391 | CL120 | HEK-293T | C41(0.94) | LDD1595 | [13] |
| LDCM0392 | CL121 | HEK-293T | C41(1.08) | LDD1596 | [13] |
| LDCM0393 | CL122 | HEK-293T | C41(1.04) | LDD1597 | [13] |
| LDCM0394 | CL123 | HEK-293T | C41(0.97) | LDD1598 | [13] |
| LDCM0395 | CL124 | HEK-293T | C41(1.04) | LDD1599 | [13] |
| LDCM0396 | CL125 | HEK-293T | C41(1.08) | LDD1600 | [13] |
| LDCM0397 | CL126 | HEK-293T | C41(1.00) | LDD1601 | [13] |
| LDCM0398 | CL127 | HEK-293T | C41(0.95) | LDD1602 | [13] |
| LDCM0399 | CL128 | HEK-293T | C41(0.97) | LDD1603 | [13] |
| LDCM0400 | CL13 | HEK-293T | C41(0.97) | LDD1604 | [13] |
| LDCM0401 | CL14 | HEK-293T | C41(0.97) | LDD1605 | [13] |
| LDCM0402 | CL15 | HEK-293T | C41(0.93) | LDD1606 | [13] |
| LDCM0403 | CL16 | HEK-293T | C41(0.96) | LDD1607 | [13] |
| LDCM0404 | CL17 | HEK-293T | C41(0.96) | LDD1608 | [13] |
| LDCM0405 | CL18 | HEK-293T | C41(0.96) | LDD1609 | [13] |
| LDCM0406 | CL19 | HEK-293T | C41(1.01) | LDD1610 | [13] |
| LDCM0407 | CL2 | HEK-293T | C41(0.97) | LDD1611 | [13] |
| LDCM0408 | CL20 | HEK-293T | C41(1.12) | LDD1612 | [13] |
| LDCM0409 | CL21 | HEK-293T | C41(0.95) | LDD1613 | [13] |
| LDCM0410 | CL22 | HEK-293T | C41(1.30) | LDD1614 | [13] |
| LDCM0411 | CL23 | HEK-293T | C41(1.30) | LDD1615 | [13] |
| LDCM0412 | CL24 | HEK-293T | C41(0.89) | LDD1616 | [13] |
| LDCM0413 | CL25 | HEK-293T | C41(1.07) | LDD1617 | [13] |
| LDCM0414 | CL26 | HEK-293T | C41(0.97) | LDD1618 | [13] |
| LDCM0415 | CL27 | HEK-293T | C41(1.00) | LDD1619 | [13] |
| LDCM0416 | CL28 | HEK-293T | C41(1.01) | LDD1620 | [13] |
| LDCM0417 | CL29 | HEK-293T | C41(0.94) | LDD1621 | [13] |
| LDCM0418 | CL3 | HEK-293T | C41(0.95) | LDD1622 | [13] |
| LDCM0419 | CL30 | HEK-293T | C41(0.91) | LDD1623 | [13] |
| LDCM0420 | CL31 | HEK-293T | C41(0.96) | LDD1624 | [13] |
| LDCM0421 | CL32 | HEK-293T | C41(1.19) | LDD1625 | [13] |
| LDCM0422 | CL33 | HEK-293T | C41(1.30) | LDD1626 | [13] |
| LDCM0423 | CL34 | HEK-293T | C41(1.19) | LDD1627 | [13] |
| LDCM0424 | CL35 | HEK-293T | C41(1.09) | LDD1628 | [13] |
| LDCM0425 | CL36 | HEK-293T | C41(0.98) | LDD1629 | [13] |
| LDCM0426 | CL37 | HEK-293T | C41(1.28) | LDD1630 | [13] |
| LDCM0428 | CL39 | HEK-293T | C41(0.91) | LDD1632 | [13] |
| LDCM0429 | CL4 | HEK-293T | C41(0.95) | LDD1633 | [13] |
| LDCM0430 | CL40 | HEK-293T | C41(1.06) | LDD1634 | [13] |
| LDCM0431 | CL41 | HEK-293T | C41(0.90) | LDD1635 | [13] |
| LDCM0432 | CL42 | HEK-293T | C41(0.98) | LDD1636 | [13] |
| LDCM0433 | CL43 | HEK-293T | C41(0.95) | LDD1637 | [13] |
| LDCM0434 | CL44 | HEK-293T | C41(1.15) | LDD1638 | [13] |
| LDCM0435 | CL45 | HEK-293T | C41(0.87) | LDD1639 | [13] |
| LDCM0436 | CL46 | HEK-293T | C41(1.22) | LDD1640 | [13] |
| LDCM0437 | CL47 | HEK-293T | C41(1.24) | LDD1641 | [13] |
| LDCM0438 | CL48 | HEK-293T | C41(1.03) | LDD1642 | [13] |
| LDCM0439 | CL49 | HEK-293T | C41(1.02) | LDD1643 | [13] |
| LDCM0440 | CL5 | HEK-293T | C41(0.99) | LDD1644 | [13] |
| LDCM0441 | CL50 | HEK-293T | C41(0.92) | LDD1645 | [13] |
| LDCM0443 | CL52 | HEK-293T | C41(0.95) | LDD1646 | [13] |
| LDCM0444 | CL53 | HEK-293T | C41(0.88) | LDD1647 | [13] |
| LDCM0445 | CL54 | HEK-293T | C41(0.95) | LDD1648 | [13] |
| LDCM0446 | CL55 | HEK-293T | C41(1.04) | LDD1649 | [13] |
| LDCM0447 | CL56 | HEK-293T | C41(1.02) | LDD1650 | [13] |
| LDCM0448 | CL57 | HEK-293T | C41(0.73) | LDD1651 | [13] |
| LDCM0449 | CL58 | HEK-293T | C41(1.21) | LDD1652 | [13] |
| LDCM0450 | CL59 | HEK-293T | C41(1.21) | LDD1653 | [13] |
| LDCM0451 | CL6 | HEK-293T | C41(1.03) | LDD1654 | [13] |
| LDCM0452 | CL60 | HEK-293T | C41(1.07) | LDD1655 | [13] |
| LDCM0453 | CL61 | HEK-293T | C41(0.90) | LDD1656 | [13] |
| LDCM0454 | CL62 | HEK-293T | C41(0.98) | LDD1657 | [13] |
| LDCM0455 | CL63 | HEK-293T | C41(0.97) | LDD1658 | [13] |
| LDCM0456 | CL64 | HEK-293T | C41(0.94) | LDD1659 | [13] |
| LDCM0457 | CL65 | HEK-293T | C41(0.90) | LDD1660 | [13] |
| LDCM0458 | CL66 | HEK-293T | C41(0.94) | LDD1661 | [13] |
| LDCM0459 | CL67 | HEK-293T | C41(0.93) | LDD1662 | [13] |
| LDCM0460 | CL68 | HEK-293T | C41(1.03) | LDD1663 | [13] |
| LDCM0461 | CL69 | HEK-293T | C41(0.87) | LDD1664 | [13] |
| LDCM0462 | CL7 | HEK-293T | C41(1.04) | LDD1665 | [13] |
| LDCM0463 | CL70 | HEK-293T | C41(1.17) | LDD1666 | [13] |
| LDCM0464 | CL71 | HEK-293T | C41(1.24) | LDD1667 | [13] |
| LDCM0465 | CL72 | HEK-293T | C41(1.05) | LDD1668 | [13] |
| LDCM0466 | CL73 | HEK-293T | C41(0.96) | LDD1669 | [13] |
| LDCM0467 | CL74 | HEK-293T | C41(1.05) | LDD1670 | [13] |
| LDCM0469 | CL76 | HEK-293T | C41(1.00) | LDD1672 | [13] |
| LDCM0470 | CL77 | HEK-293T | C41(1.04) | LDD1673 | [13] |
| LDCM0471 | CL78 | HEK-293T | C41(0.98) | LDD1674 | [13] |
| LDCM0472 | CL79 | HEK-293T | C41(1.00) | LDD1675 | [13] |
| LDCM0473 | CL8 | HEK-293T | C41(1.71) | LDD1676 | [13] |
| LDCM0474 | CL80 | HEK-293T | C41(1.24) | LDD1677 | [13] |
| LDCM0475 | CL81 | HEK-293T | C41(0.95) | LDD1678 | [13] |
| LDCM0476 | CL82 | HEK-293T | C41(1.13) | LDD1679 | [13] |
| LDCM0477 | CL83 | HEK-293T | C41(1.12) | LDD1680 | [13] |
| LDCM0478 | CL84 | HEK-293T | C41(1.15) | LDD1681 | [13] |
| LDCM0479 | CL85 | HEK-293T | C41(1.02) | LDD1682 | [13] |
| LDCM0480 | CL86 | HEK-293T | C41(1.05) | LDD1683 | [13] |
| LDCM0481 | CL87 | HEK-293T | C41(0.96) | LDD1684 | [13] |
| LDCM0482 | CL88 | HEK-293T | C41(1.01) | LDD1685 | [13] |
| LDCM0483 | CL89 | HEK-293T | C41(1.06) | LDD1686 | [13] |
| LDCM0484 | CL9 | HEK-293T | C41(0.77) | LDD1687 | [13] |
| LDCM0485 | CL90 | HEK-293T | C41(1.14) | LDD1688 | [13] |
| LDCM0486 | CL91 | HEK-293T | C41(1.00) | LDD1689 | [13] |
| LDCM0487 | CL92 | HEK-293T | C41(1.04) | LDD1690 | [13] |
| LDCM0488 | CL93 | HEK-293T | C41(1.18) | LDD1691 | [13] |
| LDCM0489 | CL94 | HEK-293T | C41(1.11) | LDD1692 | [13] |
| LDCM0490 | CL95 | HEK-293T | C41(1.10) | LDD1693 | [13] |
| LDCM0491 | CL96 | HEK-293T | C41(0.95) | LDD1694 | [13] |
| LDCM0492 | CL97 | HEK-293T | C41(1.28) | LDD1695 | [13] |
| LDCM0493 | CL98 | HEK-293T | C41(1.01) | LDD1696 | [13] |
| LDCM0494 | CL99 | HEK-293T | C41(0.92) | LDD1697 | [13] |
| LDCM0033 | Curcusone1d | MCF-7 | 2.54 | LDD0188 | [3] |
| LDCM0495 | E2913 | HEK-293T | C41(0.91) | LDD1698 | [13] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C41(14.08) | LDD1702 | [2] |
| LDCM0625 | F8 | Ramos | C41(0.98) | LDD2187 | [14] |
| LDCM0573 | Fragment11 | Ramos | C41(0.41) | LDD2190 | [14] |
| LDCM0574 | Fragment12 | Ramos | C41(2.36) | LDD2191 | [14] |
| LDCM0575 | Fragment13 | Ramos | C41(1.44) | LDD2192 | [14] |
| LDCM0576 | Fragment14 | Ramos | C41(1.21) | LDD2193 | [14] |
| LDCM0579 | Fragment20 | Ramos | C41(2.73) | LDD2194 | [14] |
| LDCM0580 | Fragment21 | Ramos | C41(0.92) | LDD2195 | [14] |
| LDCM0578 | Fragment27 | Ramos | C41(1.01) | LDD2197 | [14] |
| LDCM0586 | Fragment28 | Ramos | C41(1.50) | LDD2198 | [14] |
| LDCM0588 | Fragment30 | Ramos | C41(1.18) | LDD2199 | [14] |
| LDCM0589 | Fragment31 | Ramos | C41(1.67) | LDD2200 | [14] |
| LDCM0590 | Fragment32 | Ramos | C41(2.71) | LDD2201 | [14] |
| LDCM0468 | Fragment33 | HEK-293T | C41(0.93) | LDD1671 | [13] |
| LDCM0596 | Fragment38 | Ramos | C41(1.18) | LDD2203 | [14] |
| LDCM0427 | Fragment51 | HEK-293T | C41(0.95) | LDD1631 | [13] |
| LDCM0614 | Fragment56 | Ramos | C41(1.58) | LDD2205 | [14] |
| LDCM0569 | Fragment7 | Ramos | C41(1.45) | LDD2186 | [14] |
| LDCM0022 | KB02 | HEK-293T | C41(1.00) | LDD1492 | [13] |
| LDCM0023 | KB03 | Jurkat | C16(4.74) | LDD0209 | [5] |
| LDCM0024 | KB05 | HMCB | C41(3.85) | LDD3312 | [15] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C16(0.66) | LDD2121 | [2] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C16(0.68) | LDD2089 | [2] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C16(0.93) | LDD2093 | [2] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C16(1.08) | LDD2099 | [2] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C16(1.00) | LDD2107 | [2] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C16(0.94) | LDD2109 | [2] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C16(0.94) | LDD2111 | [2] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C16(1.84) | LDD2119 | [2] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C16(0.95) | LDD2123 | [2] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C16(0.84) | LDD2125 | [2] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C16(1.18) | LDD2127 | [2] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C16(1.20) | LDD2129 | [2] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C16(0.24) | LDD2135 | [2] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C16(1.57) | LDD2136 | [2] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C16(0.98) | LDD2137 | [2] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C16(1.38) | LDD2144 | [2] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C16(0.86) | LDD2146 | [2] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C16(0.60) | LDD2150 | [2] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C16(1.06) | LDD2207 | [16] |
The Interaction Atlas With This Target
References
