Details of the Target
General Information of Target
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K107(7.69); K113(10.00); K199(10.00); K60(6.94) | LDD0277 | [1] | |
|
DBIA Probe Info |
 |
C68(2.55) | LDD3316 | [2] | |
|
BTD Probe Info |
 |
C156(0.97) | LDD2123 | [3] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0223 | [4] | |
|
5E-2FA Probe Info |
 |
H142(0.00); H267(0.00) | LDD2235 | [5] | |
|
m-APA Probe Info |
 |
H142(0.00); H146(0.00) | LDD2231 | [5] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [6] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [6] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [6] | |
|
IPM Probe Info |
 |
N.A. | LDD0025 | [7] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [7] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [7] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [4] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [4] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [8] | |
|
HHS-482 Probe Info |
 |
Y242(0.25) | LDD2239 | [9] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C017 Probe Info |
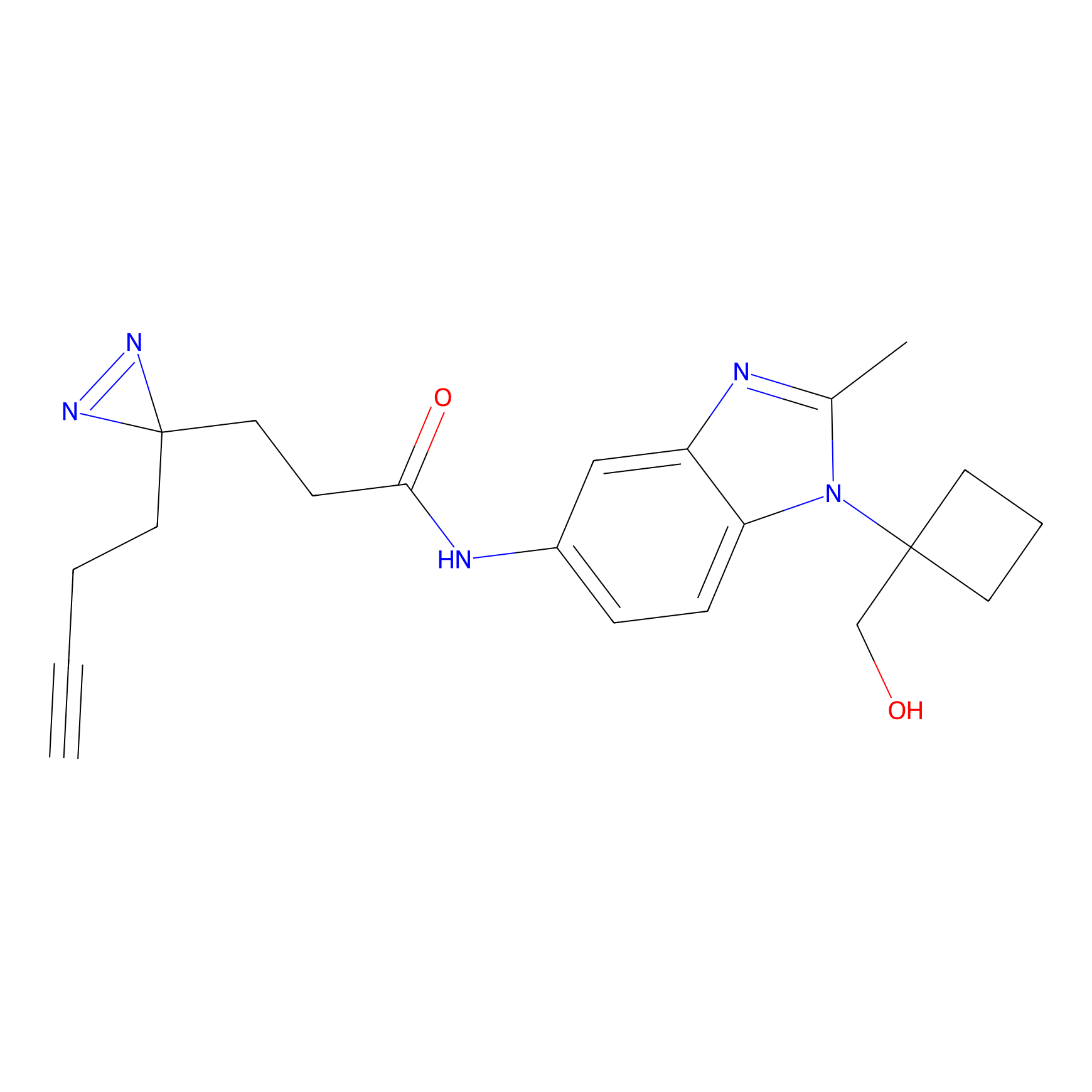 |
11.08 | LDD1725 | [10] | |
|
C026 Probe Info |
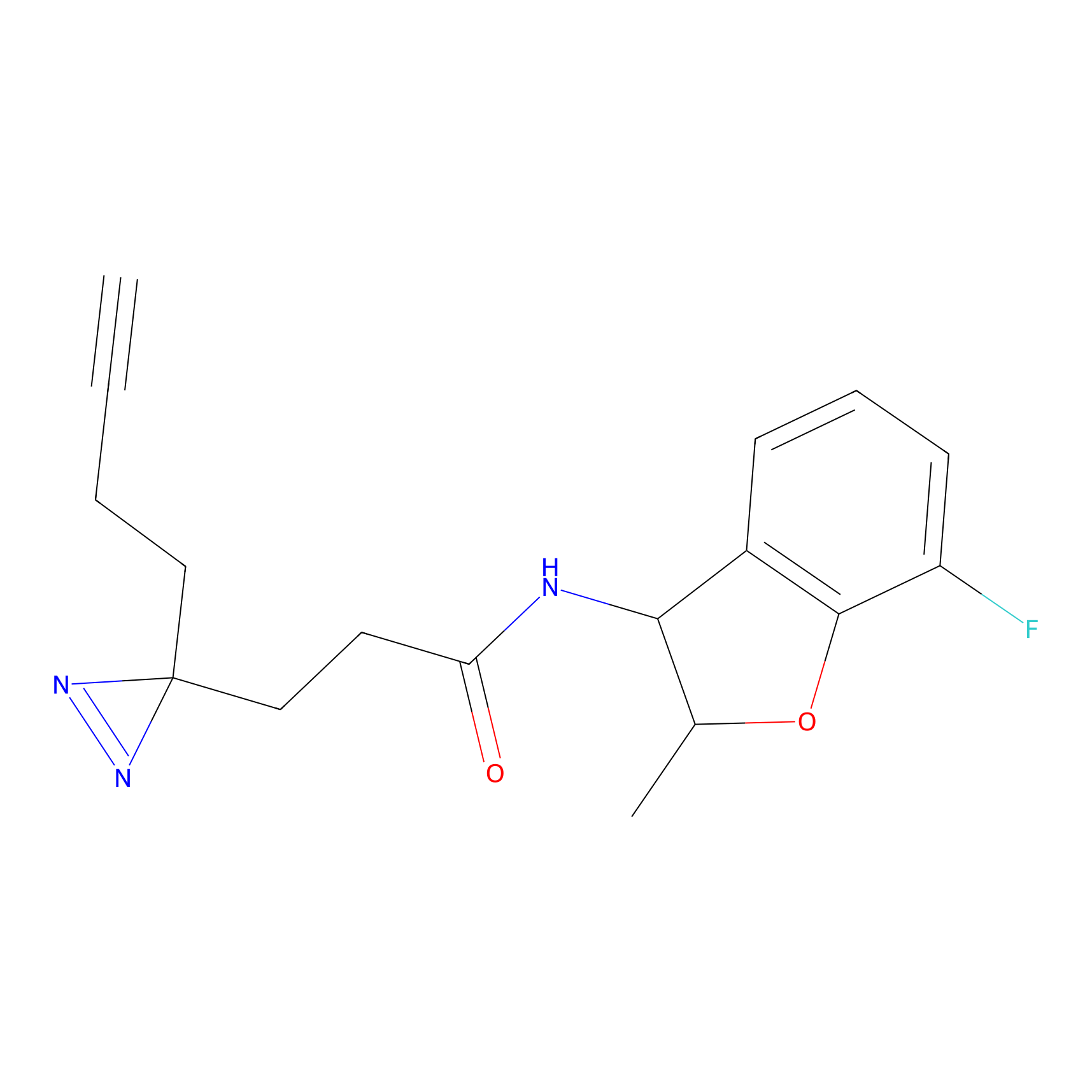 |
8.46 | LDD1732 | [10] | |
|
C092 Probe Info |
 |
23.75 | LDD1783 | [10] | |
|
C153 Probe Info |
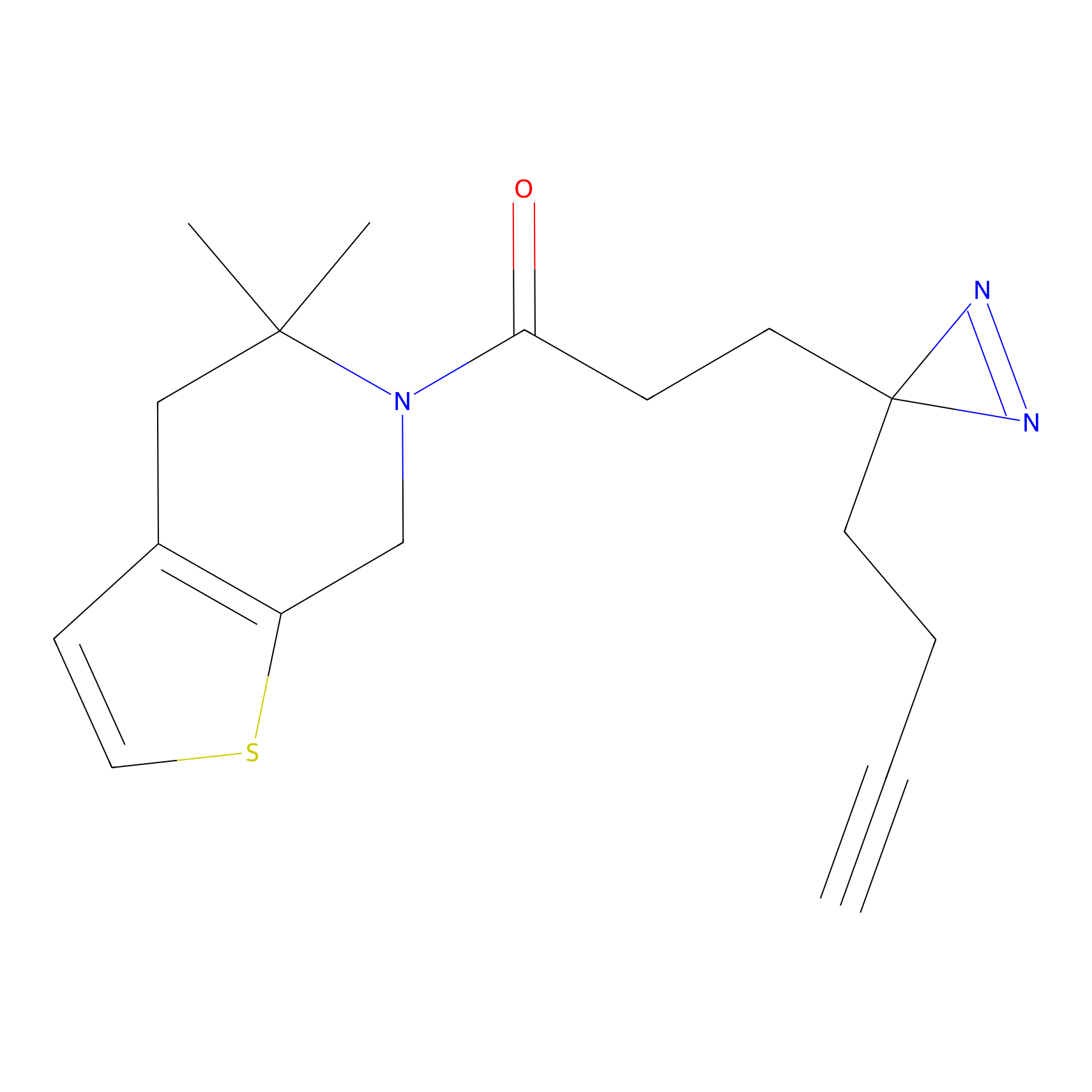 |
14.72 | LDD1834 | [10] | |
|
C178 Probe Info |
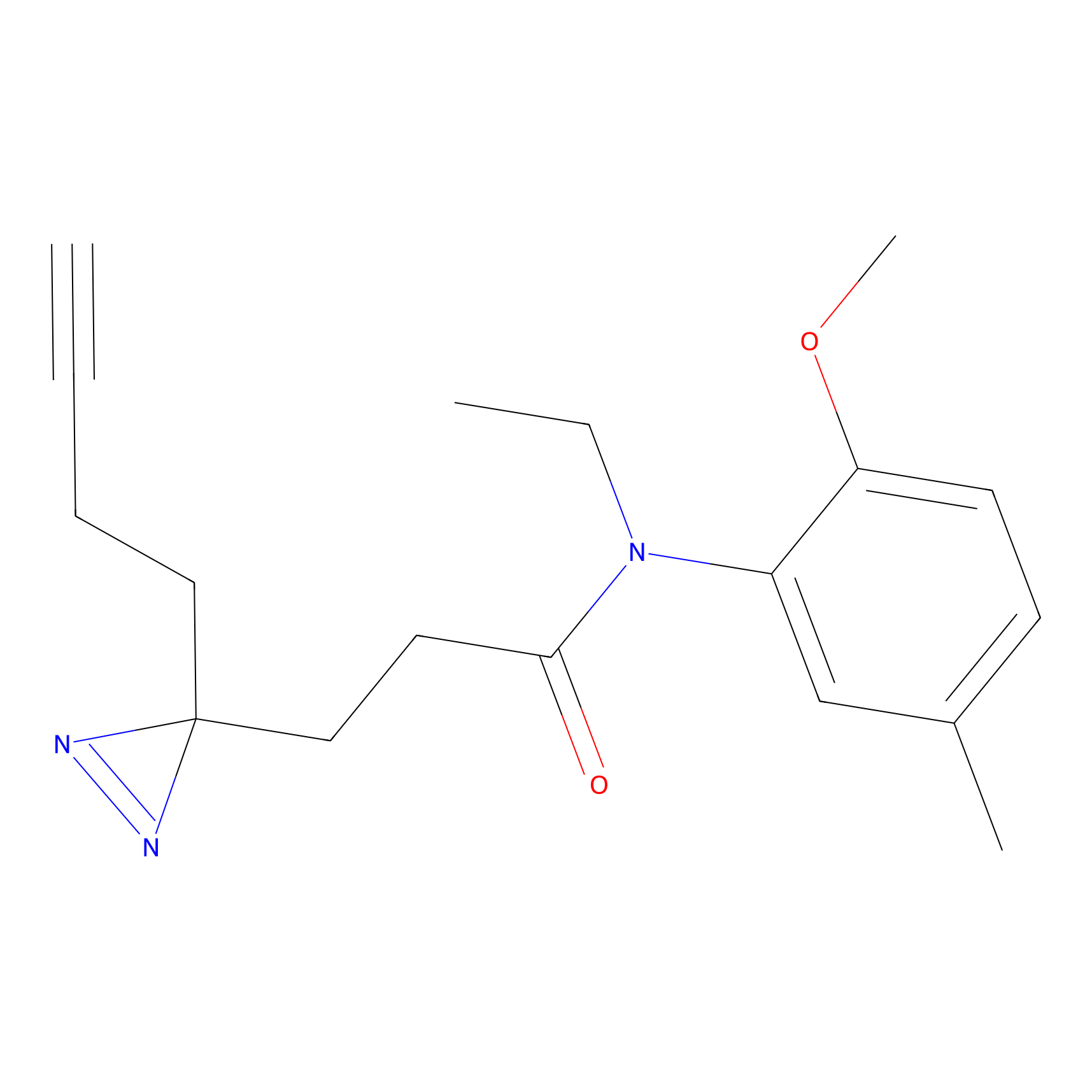 |
15.45 | LDD1857 | [10] | |
|
C388 Probe Info |
 |
42.52 | LDD2047 | [10] | |
|
C389 Probe Info |
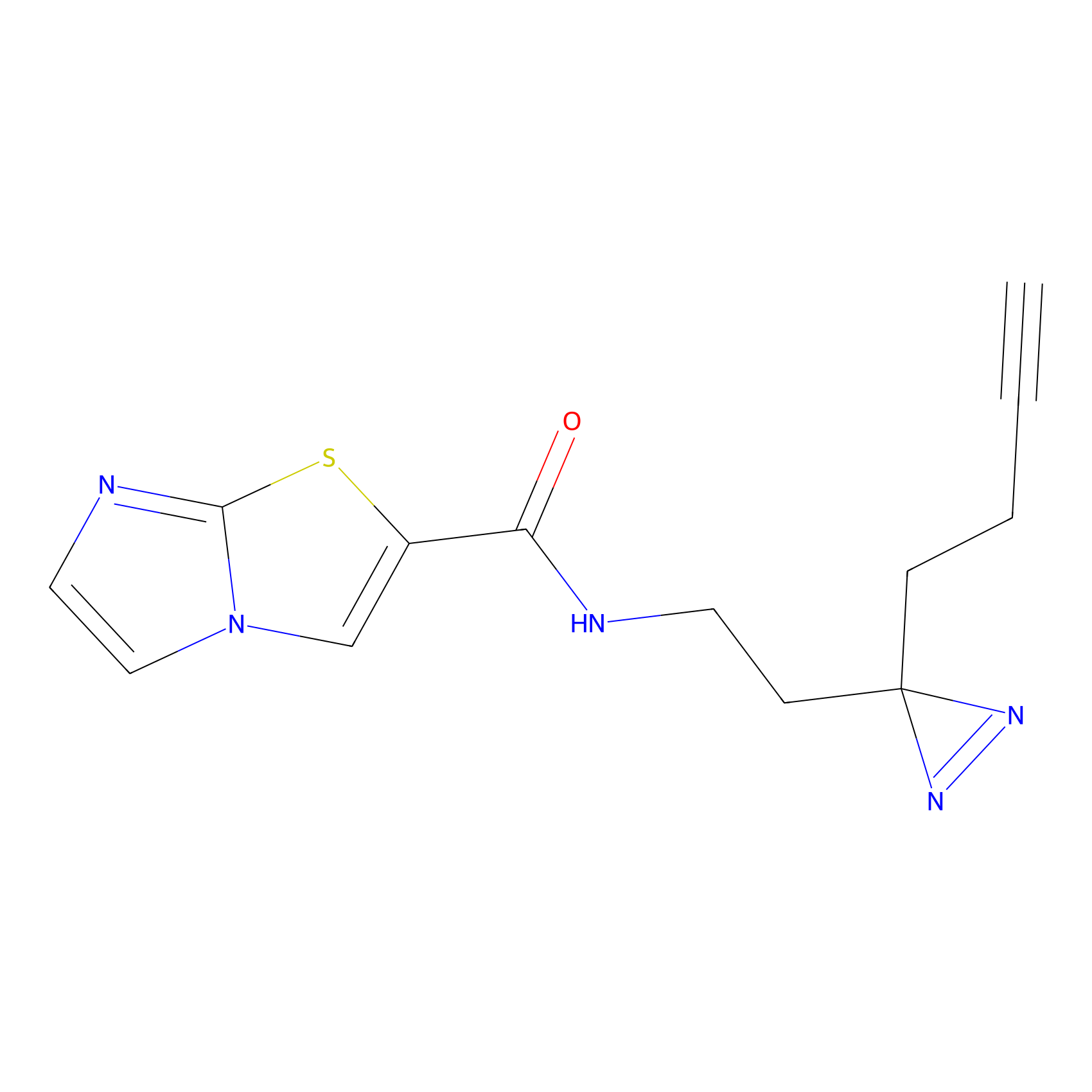 |
6.50 | LDD2048 | [10] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0226 | AC11 | HEK-293T | C290(1.10) | LDD1509 | [11] |
| LDCM0278 | AC19 | HEK-293T | C290(1.52) | LDD1517 | [11] |
| LDCM0284 | AC24 | HEK-293T | C290(0.99) | LDD1523 | [11] |
| LDCM0287 | AC27 | HEK-293T | C290(1.08) | LDD1526 | [11] |
| LDCM0290 | AC3 | HEK-293T | C290(1.03) | LDD1529 | [11] |
| LDCM0293 | AC32 | HEK-293T | C290(0.95) | LDD1532 | [11] |
| LDCM0296 | AC35 | HEK-293T | C290(1.05) | LDD1535 | [11] |
| LDCM0302 | AC40 | HEK-293T | C290(1.03) | LDD1541 | [11] |
| LDCM0305 | AC43 | HEK-293T | C290(1.09) | LDD1544 | [11] |
| LDCM0310 | AC48 | HEK-293T | C290(1.04) | LDD1549 | [11] |
| LDCM0314 | AC51 | HEK-293T | C290(1.00) | LDD1553 | [11] |
| LDCM0319 | AC56 | HEK-293T | C290(0.99) | LDD1558 | [11] |
| LDCM0322 | AC59 | HEK-293T | C290(1.08) | LDD1561 | [11] |
| LDCM0328 | AC64 | HEK-293T | C290(1.00) | LDD1567 | [11] |
| LDCM0345 | AC8 | HEK-293T | C290(0.98) | LDD1569 | [11] |
| LDCM0275 | AKOS034007705 | HEK-293T | C290(1.01) | LDD1514 | [11] |
| LDCM0369 | CL100 | HEK-293T | C290(1.12) | LDD1573 | [11] |
| LDCM0373 | CL104 | HEK-293T | C290(1.03) | LDD1577 | [11] |
| LDCM0377 | CL108 | HEK-293T | C290(0.98) | LDD1581 | [11] |
| LDCM0382 | CL112 | HEK-293T | C290(1.03) | LDD1586 | [11] |
| LDCM0386 | CL116 | HEK-293T | C290(1.04) | LDD1590 | [11] |
| LDCM0390 | CL12 | HEK-293T | C290(1.09) | LDD1594 | [11] |
| LDCM0391 | CL120 | HEK-293T | C290(1.07) | LDD1595 | [11] |
| LDCM0395 | CL124 | HEK-293T | C290(1.05) | LDD1599 | [11] |
| LDCM0399 | CL128 | HEK-293T | C290(1.05) | LDD1603 | [11] |
| LDCM0403 | CL16 | HEK-293T | C290(1.07) | LDD1607 | [11] |
| LDCM0406 | CL19 | HEK-293T | C290(1.09) | LDD1610 | [11] |
| LDCM0412 | CL24 | HEK-293T | C290(1.04) | LDD1616 | [11] |
| LDCM0416 | CL28 | HEK-293T | C290(1.00) | LDD1620 | [11] |
| LDCM0420 | CL31 | HEK-293T | C290(1.11) | LDD1624 | [11] |
| LDCM0425 | CL36 | HEK-293T | C290(0.97) | LDD1629 | [11] |
| LDCM0429 | CL4 | HEK-293T | C290(1.13) | LDD1633 | [11] |
| LDCM0430 | CL40 | HEK-293T | C290(1.06) | LDD1634 | [11] |
| LDCM0433 | CL43 | HEK-293T | C290(0.98) | LDD1637 | [11] |
| LDCM0438 | CL48 | HEK-293T | C290(1.00) | LDD1642 | [11] |
| LDCM0443 | CL52 | HEK-293T | C290(1.09) | LDD1646 | [11] |
| LDCM0446 | CL55 | HEK-293T | C290(1.00) | LDD1649 | [11] |
| LDCM0452 | CL60 | HEK-293T | C290(1.13) | LDD1655 | [11] |
| LDCM0456 | CL64 | HEK-293T | C290(1.02) | LDD1659 | [11] |
| LDCM0459 | CL67 | HEK-293T | C290(1.16) | LDD1662 | [11] |
| LDCM0462 | CL7 | HEK-293T | C290(1.01) | LDD1665 | [11] |
| LDCM0465 | CL72 | HEK-293T | C290(1.01) | LDD1668 | [11] |
| LDCM0469 | CL76 | HEK-293T | C290(1.09) | LDD1672 | [11] |
| LDCM0472 | CL79 | HEK-293T | C290(1.19) | LDD1675 | [11] |
| LDCM0478 | CL84 | HEK-293T | C290(1.02) | LDD1681 | [11] |
| LDCM0482 | CL88 | HEK-293T | C290(1.03) | LDD1685 | [11] |
| LDCM0486 | CL91 | HEK-293T | C290(1.08) | LDD1689 | [11] |
| LDCM0491 | CL96 | HEK-293T | C290(1.12) | LDD1694 | [11] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C156(1.58) | LDD1702 | [3] |
| LDCM0625 | F8 | Ramos | C290(1.94) | LDD2187 | [12] |
| LDCM0573 | Fragment11 | Ramos | C290(2.80) | LDD2190 | [12] |
| LDCM0574 | Fragment12 | Ramos | C290(0.78) | LDD2191 | [12] |
| LDCM0576 | Fragment14 | Ramos | C290(1.04) | LDD2193 | [12] |
| LDCM0580 | Fragment21 | Ramos | C290(1.48) | LDD2195 | [12] |
| LDCM0582 | Fragment23 | Ramos | C290(0.98) | LDD2196 | [12] |
| LDCM0578 | Fragment27 | Ramos | C290(0.75) | LDD2197 | [12] |
| LDCM0588 | Fragment30 | Ramos | C290(2.38) | LDD2199 | [12] |
| LDCM0589 | Fragment31 | Ramos | C290(0.93) | LDD2200 | [12] |
| LDCM0566 | Fragment4 | Ramos | C290(0.99) | LDD2184 | [12] |
| LDCM0614 | Fragment56 | Ramos | C290(1.21) | LDD2205 | [12] |
| LDCM0569 | Fragment7 | Ramos | C290(1.22) | LDD2186 | [12] |
| LDCM0022 | KB02 | 8305C | C68(1.28) | LDD2248 | [2] |
| LDCM0023 | KB03 | Ramos | C290(1.32) | LDD2183 | [12] |
| LDCM0024 | KB05 | MEL167 | C68(2.55) | LDD3316 | [2] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [4] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C156(0.97) | LDD2123 | [3] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Huntingtin (HTT) | Huntingtin family | P42858 | |||
| Wolframin (WFS1) | . | O76024 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Progranulin (GRN) | Granulin family | P28799 | |||
| Heat shock protein beta-1 (HSPB1) | Small heat shock protein (HSP20) family | P04792 | |||
References
