Details of the Target
General Information of Target
| Target ID | LDTP07221 | |||||
|---|---|---|---|---|---|---|
| Target Name | Myomegalin (PDE4DIP) | |||||
| Gene Name | PDE4DIP | |||||
| Gene ID | 9659 | |||||
| Synonyms |
CMYA2; KIAA0454; KIAA0477; MMGL; Myomegalin; Cardiomyopathy-associated protein 2; Phosphodiesterase 4D-interacting protein |
|||||
| 3D Structure | ||||||
| Sequence |
MSNGYRTLSQHLNDLKKENFSLKLRIYFLEERMQQKYEASREDIYKRNIELKVEVESLKR
ELQDKKQHLDKTWADVENLNSQNEAELRRQFEERQQETEHVYELLENKIQLLQEESRLAK NEAARMAALVEAEKECNLELSEKLKGVTKNWEDVPGDQVKPDQYTEALAQRDKRIEELNQ SLAAQERLVEQLSREKQQLLHLLEEPTSMEVQPMTEELLKQQKLNSHETTITQQSVSDSH LAELQEKIQQTEATNKILQEKLNEMSYELKCAQESSQKQDGTIQNLKETLKSRERETEEL YQVIEGQNDTMAKLREMLHQSQLGQLHSSEGTSPAQQQVALLDLQSALFCSQLEIQKLQR VVRQKERQLADAKQCVQFVEAAAHESEQQKEASWKHNQELRKALQQLQEELQNKSQQLRA WEAEKYNEIRTQEQNIQHLNHSLSHKEQLLQEFRELLQYRDNSDKTLEANEMLLEKLRQR IHDKAVALERAIDEKFSALEEKEKELRQLRLAVRERDHDLERLRDVLSSNEATMQSMESL LRAKGLEVEQLSTTCQNLQWLKEEMETKFSRWQKEQESIIQQLQTSLHDRNKEVEDLSAT LLCKLGPGQSEIAEELCQRLQRKERMLQDLLSDRNKQVLEHEMEIQGLLQSVSTREQESQ AAAEKLVQALMERNSELQALRQYLGGRDSLMSQAPISNQQAEVTPTGRLGKQTDQGSMQI PSRDDSTSLTAKEDVSIPRSTLGDLDTVAGLEKELSNAKEELELMAKKERESQMELSALQ SMMAVQEEELQVQAADMESLTRNIQIKEDLIKDLQMQLVDPEDIPAMERLTQEVLLLREK VASVESQGQEISGNRRQQLLLMLEGLVDERSRLNEALQAERQLYSSLVKFHAHPESSERD RTLQVELEGAQVLRSRLEEVLGRSLERLNRLETLAAIGGAAAGDDTEDTSTEFTDSIEEE AAHHSHQQLVKVALEKSLATVETQNPSFSPPSPMGGDSNRCLQEEMLHLRAEFHQHLEEK RKAEEELKELKAQIEEAGFSSVSHIRNTMLSLCLENAELKEQMGEAMSDGWEIEEDKEKG EVMVETVVTKEGLSESSLQAEFRKLQGKLKNAHNIINLLKEQLVLSSKEGNSKLTPELLV HLTSTIERINTELVGSPGKHQHQEEGNVTVRPFPRPQSLDLGATFTVDAHQLDNQSQPRD PGPQSAFSLPGSTQHLRSQLSQCKQRYQDLQEKLLLSEATVFAQANELEKYRVMLTGESL VKQDSKQIQVDLQDLGYETCGRSENEAEREETTSPECEEHNSLKEMVLMEGLCSEQGRRG STLASSSERKPLENQLGKQEEFRVYGKSENILVLRKDIKDLKAQLQNANKVIQNLKSRVR SLSVTSDYSSSLERPRKLRAVGTLEGSSPHSVPDEDEGWLSDGTGAFYSPGLQAKKDLES LIQRVSQLEAQLPKNGLEEKLAEELRSASWPGKYDSLIQDQARELSYLRQKIREGRGICY LITRHAKDTVKSFEDLLRSNDIDYYLGQSFREQLAQGSQLTERLTSKLSTKDHKSEKDQA GLEPLALRLSRELQEKEKVIEVLQAKLDARSLTPSSSHALSDSHRSPSSTSFLSDELEAC SDMDIVSEYTHYEEKKASPSHSDSIHHSSHSAVLSSKPSSTSASQGAKAESNSNPISLPT PQNTPKEANQAHSGFHFHSIPKLASLPQAPLPSAPSSFLPFSPTGPLLLGCCETPVVSLA EAQQELQMLQKQLGESASTVPPASTATLLSNDLEADSSYYLNSAQPHSPPRGTIELGRIL EPGYLGSSGKWDVMRPQKGSVSGDLSSGSSVYQLNSKPTGADLLEEHLGEIRNLRQRLEE SICINDRLREQLEHRLTSTARGRGSTSNFYSQGLESIPQLCNENRVLREDNRRLQAQLSH VSREHSQETESLREALLSSRSHLQELEKELEHQKVERQQLLEDLREKQQEVLHFREERLS LQENDSRLQHKLVLLQQQCEEKQQLFESLQSELQIYEALYGNSKKGLKAYSLDACHQIPL SSDLSHLVAEVRALRGQLEQSIQGNNCLRLQLQQQLESGAGKASLSPSSINQNFPASTDP GNKQLLLQDSAVSPPVRDVGMNSPALVFPSSASSTPGSETPIINRANGLGLDTSPVMKTP PKLEGDATDGSFANKHGRHVIGHIDDYSALRQQIAEGKLLVKKIVSLVRSACSFPGLEAQ GTEVLGSKGIHELRSSTSALHHALEESASLLTMFWRAALPSTHIPVLPGKVGESTERELL ELRTKVSKQERLLQSTTEHLKNANQQKESMEQFIVSQLTRTHDVLKKARTNLEVKSLRAL PCTPAL |
|||||
| Target Bioclass |
Other
|
|||||
| Subcellular location |
Cytoplasm, cytoskeleton, microtubule organizing center, centrosome; Golgi apparatus
|
|||||
| Function |
Functions as an anchor sequestering components of the cAMP-dependent pathway to Golgi and/or centrosomes.; [Isoform 13]: Participates in microtubule dynamics, promoting microtubule assembly. Depending upon the cell context, may act at the level of the Golgi apparatus or that of the centrosome. In complex with AKAP9, recruits CAMSAP2 to the Golgi apparatus and tethers non-centrosomal minus-end microtubules to the Golgi, an important step for polarized cell movement. In complex with AKAP9, EB1/MAPRE1 and CDK5RAP2, contributes to microtubules nucleation and extension from the centrosome to the cell periphery, a crucial process for directed cell migration, mitotic spindle orientation and cell-cycle progression.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K1362(3.85); K484(9.37); K592(10.00) | LDD0277 | [1] | |
|
DBIA Probe Info |
 |
C434(1.34) | LDD3311 | [2] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0162 | [3] | |
|
Lodoacetamide azide Probe Info |
 |
C375(0.00); C271(0.00) | LDD0037 | [4] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [5] | |
|
PF-06672131 Probe Info |
 |
N.A. | LDD0017 | [6] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [7] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [5] | |
|
NAIA_5 Probe Info |
 |
C271(0.00); C603(0.00) | LDD2223 | [8] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C186 Probe Info |
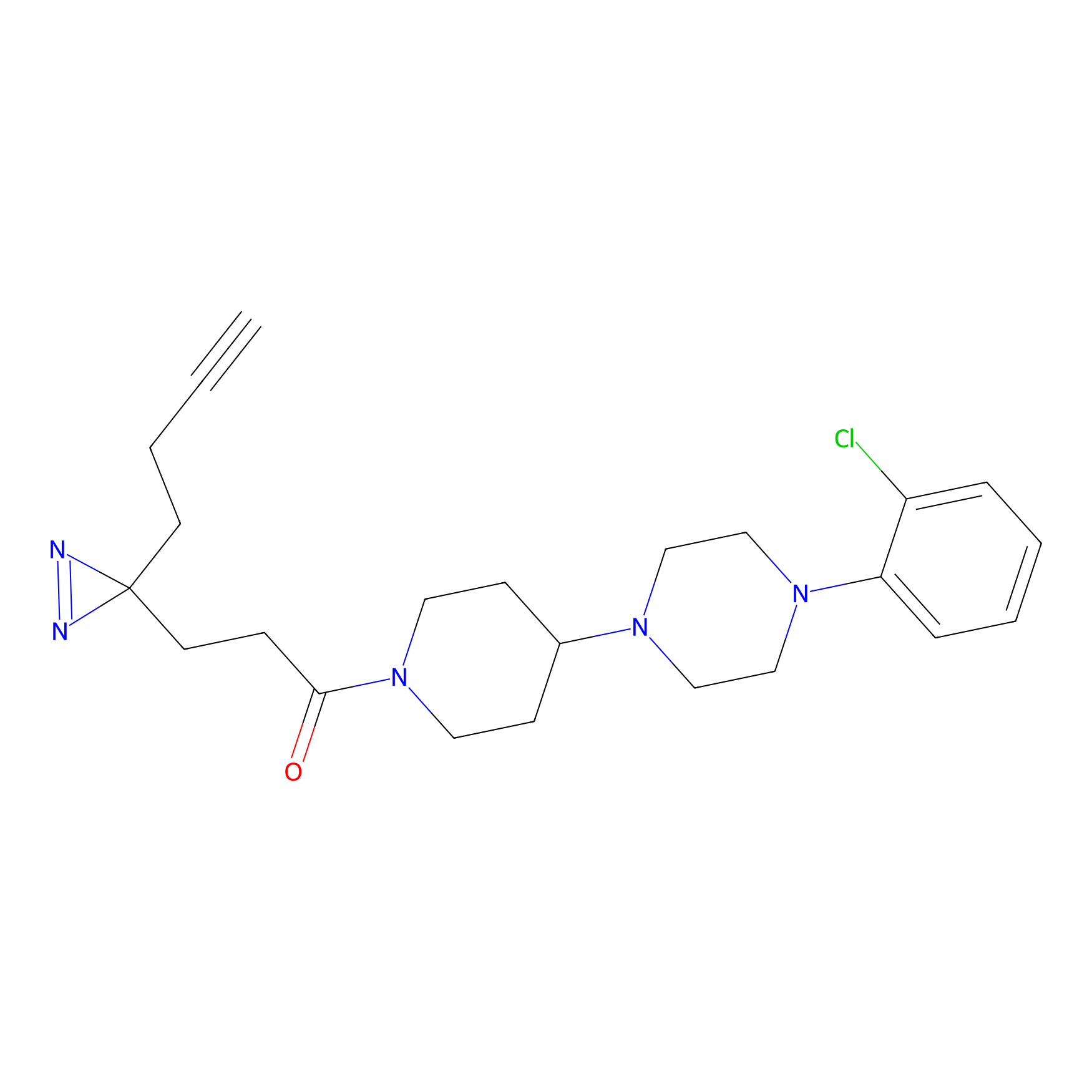 |
8.34 | LDD1864 | [9] | |
|
C191 Probe Info |
 |
23.59 | LDD1868 | [9] | |
|
C269 Probe Info |
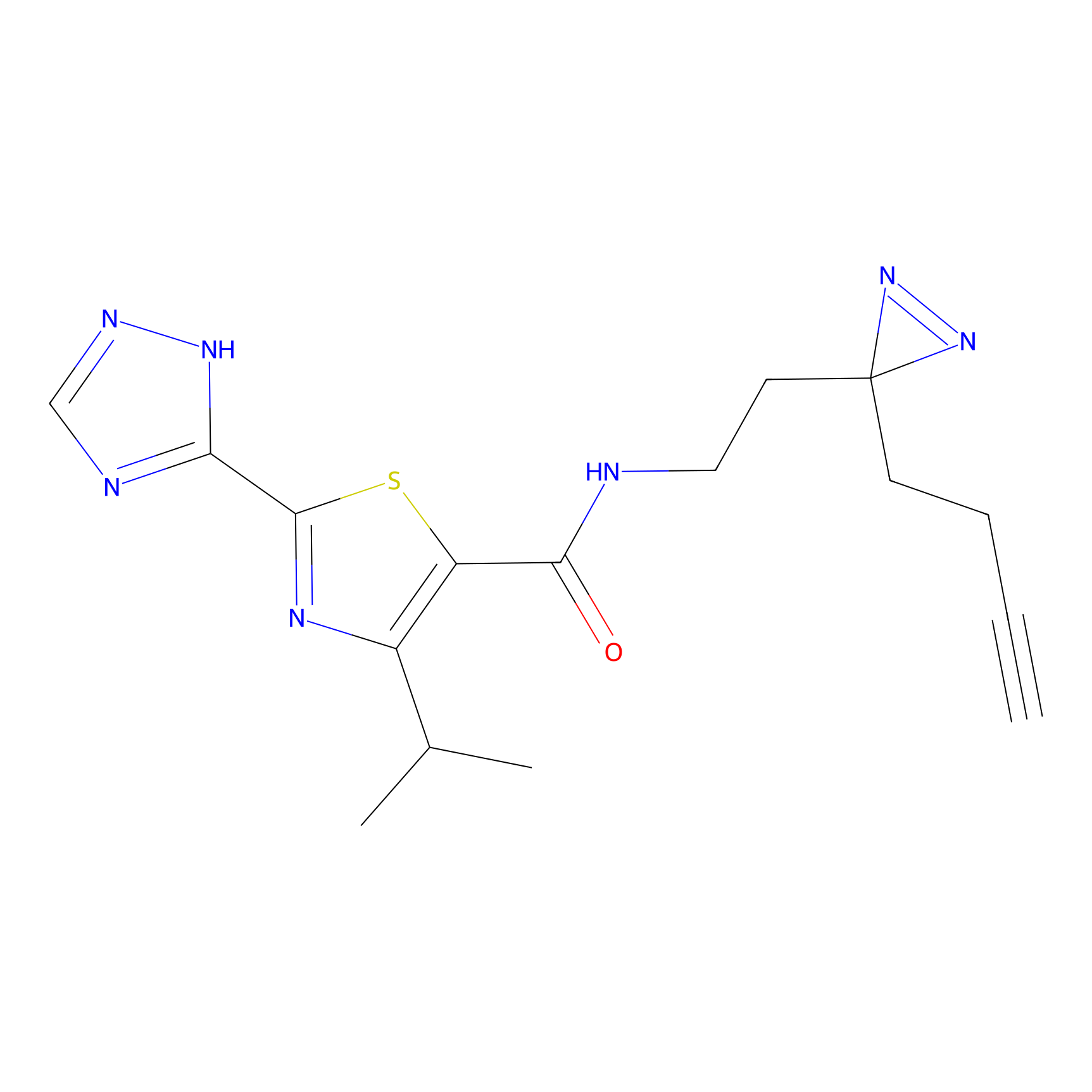 |
5.94 | LDD1939 | [9] | |
|
C407 Probe Info |
 |
14.72 | LDD2064 | [9] | |
|
C429 Probe Info |
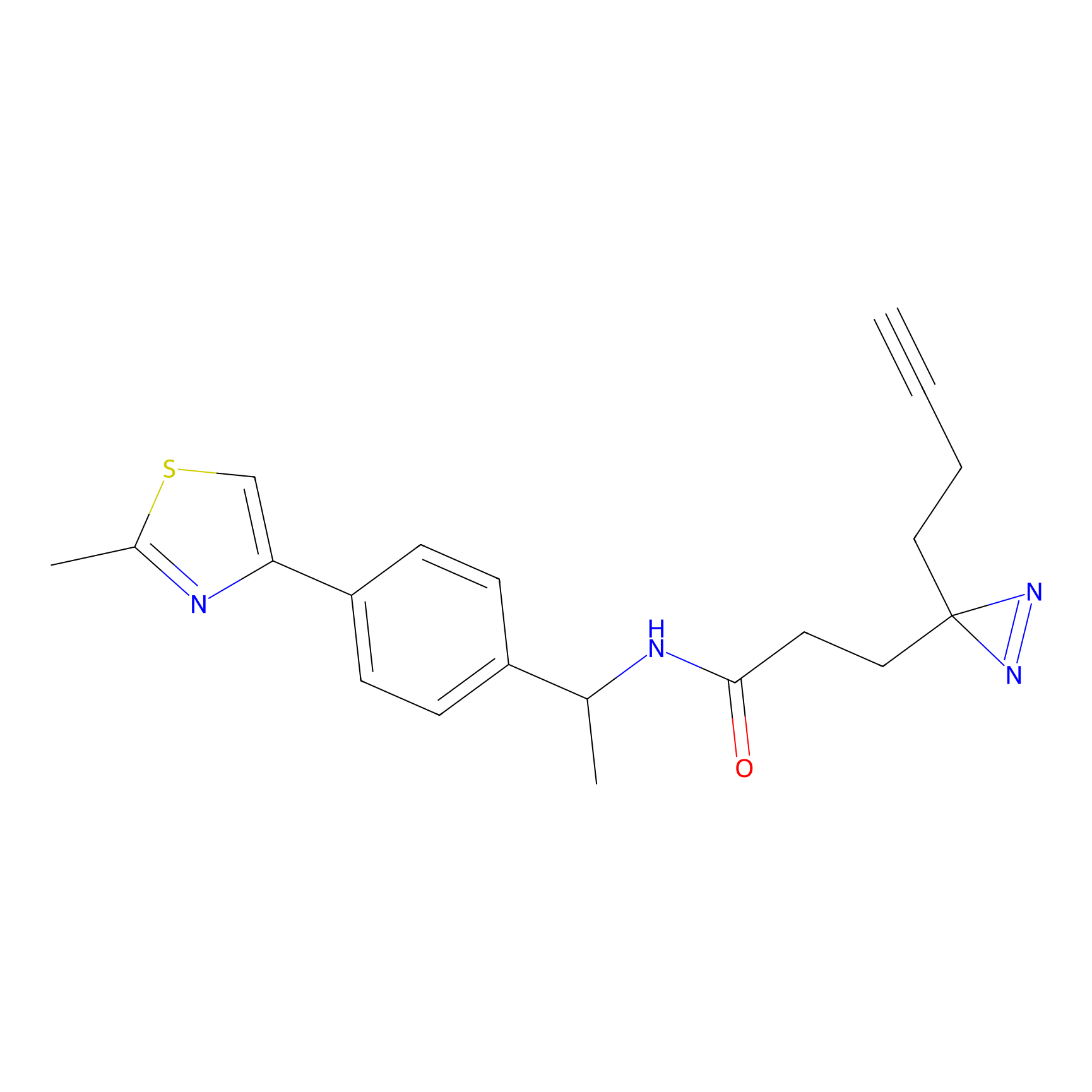 |
14.93 | LDD2084 | [9] | |
|
C433 Probe Info |
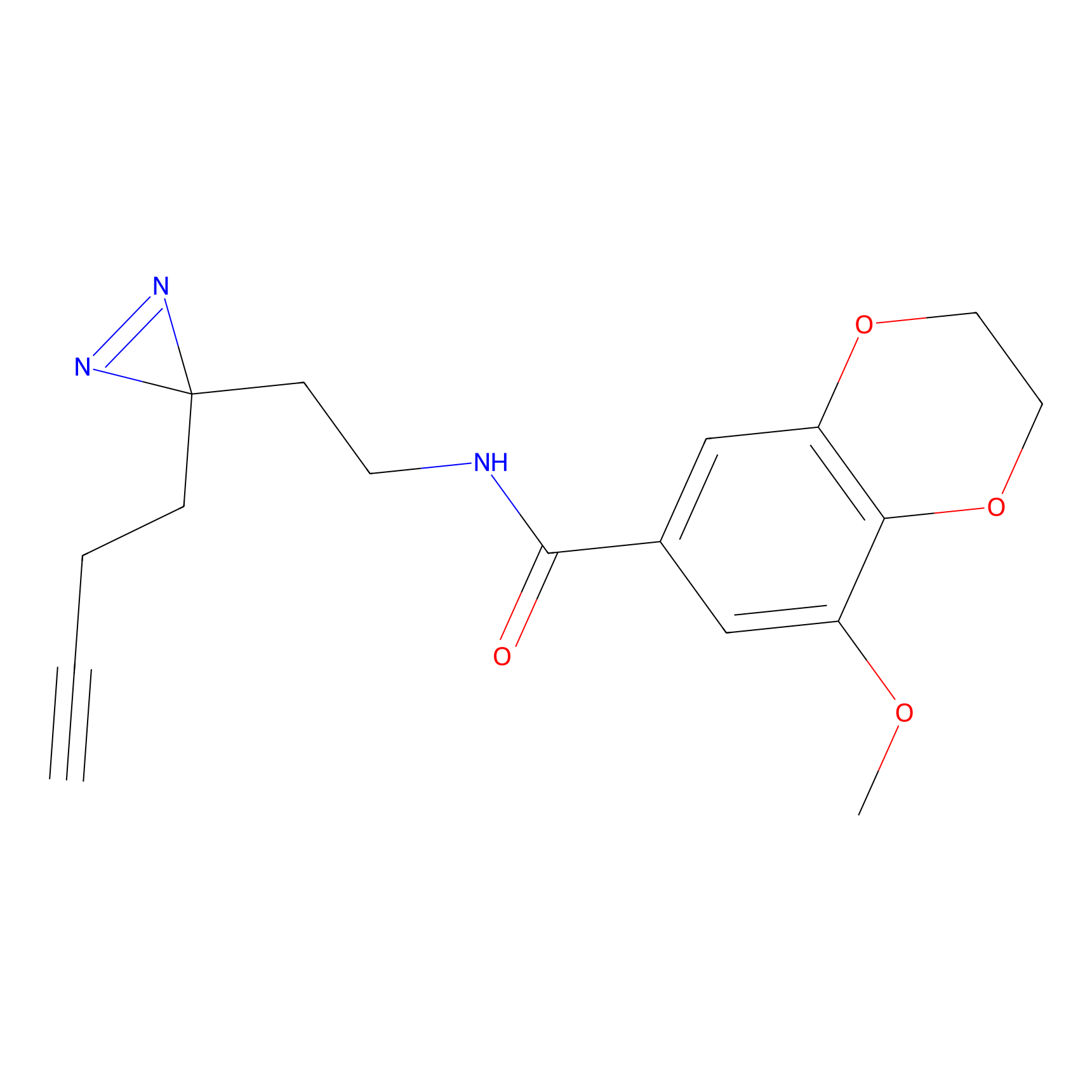 |
6.36 | LDD2088 | [9] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0139 | [10] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0561 | Abegg_cp(-)-10 | HeLa | C617(2.14) | LDD0312 | [7] |
| LDCM0632 | CL-Sc | Hep-G2 | C271(2.86); C603(2.20); C375(1.89); C603(1.04) | LDD2227 | [8] |
| LDCM0022 | KB02 | 22RV1 | C434(1.24); C780(1.74) | LDD2243 | [2] |
| LDCM0023 | KB03 | 22RV1 | C434(1.28); C780(1.99) | LDD2660 | [2] |
| LDCM0024 | KB05 | G361 | C434(1.34) | LDD3311 | [2] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| MICOS complex subunit MIC60 (IMMT) | MICOS complex subunit Mic60 family | Q16891 | |||
| Sodium channel modifier 1 (SCNM1) | . | Q9BWG6 | |||
| Syntenin-1 (SDCBP) | . | O00560 | |||
Transcription factor
Immunoglobulin
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Myosin-binding protein C, cardiac-type (MYBPC3) | MyBP family | Q14896 | |||
Other
References
