Details of the Target
General Information of Target
| Target ID | LDTP00252 | |||||
|---|---|---|---|---|---|---|
| Target Name | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform (PIK3CD) | |||||
| Gene Name | PIK3CD | |||||
| Gene ID | 5293 | |||||
| Synonyms |
Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform; PI3-kinase subunit delta; PI3K-delta; PI3Kdelta; PtdIns-3-kinase subunit delta; EC 2.7.1.137; EC 2.7.1.153; Phosphatidylinositol 4,5-bisphosphate 3-kinase 110 kDa catalytic subunit delta; PtdIns-3-kinase subunit p110-delta; p110delta
|
|||||
| 3D Structure | ||||||
| Sequence |
MPPGVDCPMEFWTKEENQSVVVDFLLPTGVYLNFPVSRNANLSTIKQLLWHRAQYEPLFH
MLSGPEAYVFTCINQTAEQQELEDEQRRLCDVQPFLPVLRLVAREGDRVKKLINSQISLL IGKGLHEFDSLCDPEVNDFRAKMCQFCEEAAARRQQLGWEAWLQYSFPLQLEPSAQTWGP GTLRLPNRALLVNVKFEGSEESFTFQVSTKDVPLALMACALRKKATVFRQPLVEQPEDYT LQVNGRHEYLYGSYPLCQFQYICSCLHSGLTPHLTMVHSSSILAMRDEQSNPAPQVQKPR AKPPPIPAKKPSSVSLWSLEQPFRIELIQGSKVNADERMKLVVQAGLFHGNEMLCKTVSS SEVSVCSEPVWKQRLEFDINICDLPRMARLCFALYAVIEKAKKARSTKKKSKKADCPIAW ANLMLFDYKDQLKTGERCLYMWPSVPDEKGELLNPTGTVRSNPNTDSAAALLICLPEVAP HPVYYPALEKILELGRHSECVHVTEEEQLQLREILERRGSGELYEHEKDLVWKLRHEVQE HFPEALARLLLVTKWNKHEDVAQMLYLLCSWPELPVLSALELLDFSFPDCHVGSFAIKSL RKLTDDELFQYLLQLVQVLKYESYLDCELTKFLLDRALANRKIGHFLFWHLRSEMHVPSV ALRFGLILEAYCRGSTHHMKVLMKQGEALSKLKALNDFVKLSSQKTPKPQTKELMHLCMR QEAYLEALSHLQSPLDPSTLLAEVCVEQCTFMDSKMKPLWIMYSNEEAGSGGSVGIIFKN GDDLRQDMLTLQMIQLMDVLWKQEGLDLRMTPYGCLPTGDRTGLIEVVLRSDTIANIQLN KSNMAATAAFNKDALLNWLKSKNPGEALDRAIEEFTLSCAGYCVATYVLGIGDRHSDNIM IRESGQLFHIDFGHFLGNFKTKFGINRERVPFILTYDFVHVIQQGKTNNSEKFERFRGYC ERAYTILRRHGLLFLHLFALMRAAGLPELSCSKDIQYLKDSLALGKTEEEALKHFRVKFN EALRESWKTKVNWLAHNVSKDNRQ |
|||||
| Target Type |
Successful
|
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
PI3/PI4-kinase family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Phosphoinositide-3-kinase (PI3K) phosphorylates phosphatidylinositol (PI) and its phosphorylated derivatives at position 3 of the inositol ring to produce 3-phosphoinositides. Uses ATP and PtdIns(4,5)P2 (phosphatidylinositol 4,5-bisphosphate) to generate phosphatidylinositol 3,4,5-trisphosphate (PIP3). PIP3 plays a key role by recruiting PH domain-containing proteins to the membrane, including AKT1 and PDPK1, activating signaling cascades involved in cell growth, survival, proliferation, motility and morphology. Mediates immune responses. Plays a role in B-cell development, proliferation, migration, and function. Required for B-cell receptor (BCR) signaling. Mediates B-cell proliferation response to anti-IgM, anti-CD40 and IL4 stimulation. Promotes cytokine production in response to TLR4 and TLR9. Required for antibody class switch mediated by TLR9. Involved in the antigen presentation function of B-cells. Involved in B-cell chemotaxis in response to CXCL13 and sphingosine 1-phosphate (S1P). Required for proliferation, signaling and cytokine production of naive, effector and memory T-cells. Required for T-cell receptor (TCR) signaling. Mediates TCR signaling events at the immune synapse. Activation by TCR leads to antigen-dependent memory T-cell migration and retention to antigenic tissues. Together with PIK3CG participates in T-cell development. Contributes to T-helper cell expansion and differentiation. Required for T-cell migration mediated by homing receptors SELL/CD62L, CCR7 and S1PR1 and antigen dependent recruitment of T-cells. Together with PIK3CG is involved in natural killer (NK) cell development and migration towards the sites of inflammation. Participates in NK cell receptor activation. Plays a role in NK cell maturation and cytokine production. Together with PIK3CG is involved in neutrophil chemotaxis and extravasation. Together with PIK3CG participates in neutrophil respiratory burst. Plays important roles in mast-cell development and mast cell mediated allergic response. Involved in stem cell factor (SCF)-mediated proliferation, adhesion and migration. Required for allergen-IgE-induced degranulation and cytokine release. The lipid kinase activity is required for its biological function. Isoform 2 may be involved in stabilizing total RAS levels, resulting in increased ERK phosphorylation and increased PI3K activity.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| HEC1 | SNV: p.S764N | DBIA Probe Info | |||
| HEC1B | SNV: p.S764N | . | |||
| HT115 | SNV: p.L725I; p.E1021K | . | |||
| Ishikawa (Heraklio) 02 ER | Deletion: p.N464TfsTer83 | . | |||
| JURKAT | SNV: p.A396T | . | |||
| MDAMB453 | SNV: p.G4E | . | |||
| MEWO | SNV: p.P186S | DBIA Probe Info | |||
| MOLT4 | SNV: p.A388T; p.S738N | IA-alkyne Probe Info | |||
| NCIH1993 | SNV: p.E320D; p.E320Ter | . | |||
| PEO1 | SNV: p.C132Y | . | |||
| REH | SNV: p.E1021K | DBIA Probe Info | |||
| SNU1 | SNV: p.P294H | DBIA Probe Info | |||
| TE8 | SNV: p.M683I | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
BTD Probe Info |
 |
C991(0.56) | LDD2140 | [1] | |
|
Johansson_61 Probe Info |
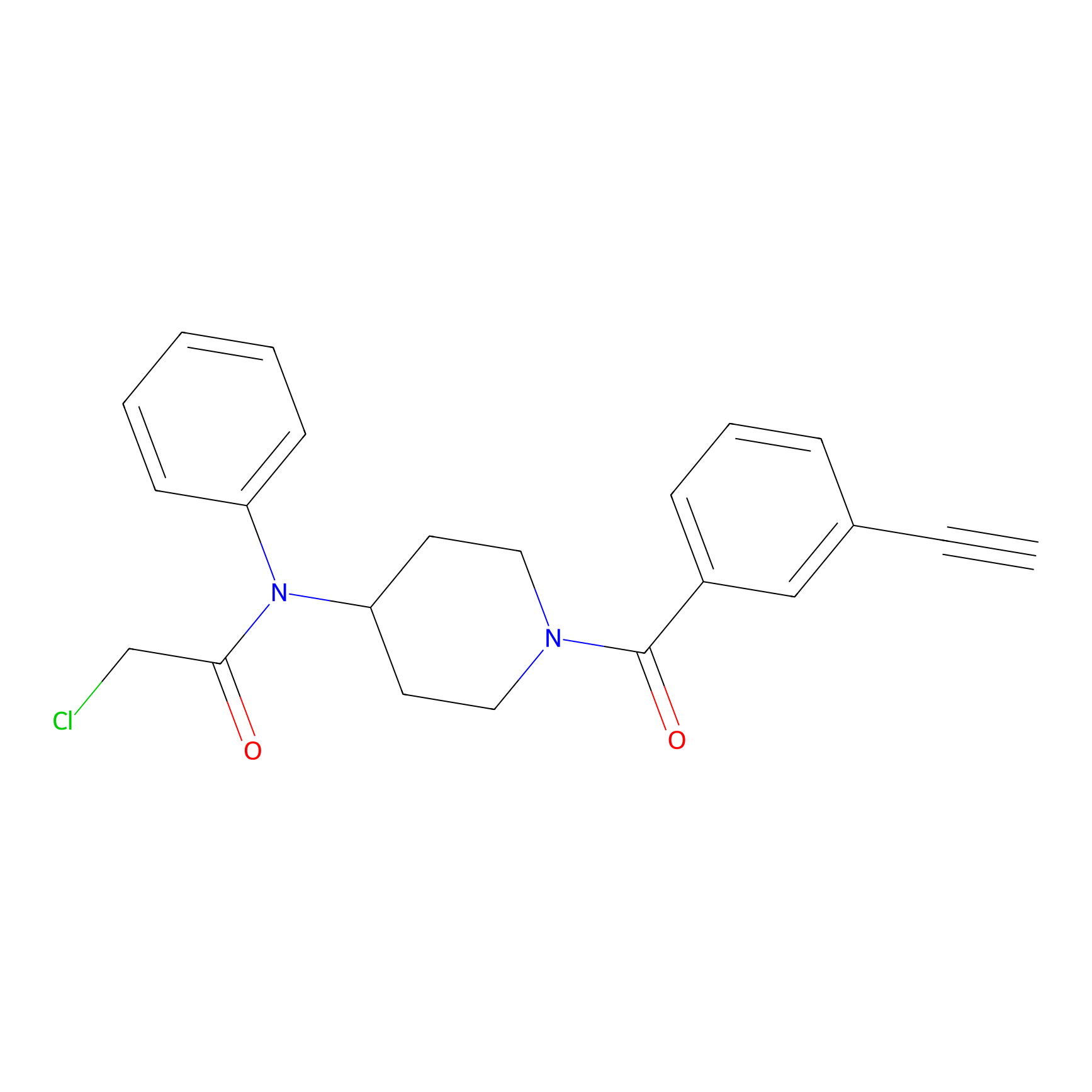 |
_(6.74) | LDD1485 | [2] | |
|
AHL-Pu-1 Probe Info |
 |
C474(3.29) | LDD0170 | [3] | |
|
DBIA Probe Info |
 |
C991(6.96) | LDD0209 | [4] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C991(0.00); C219(0.00); C132(0.00); C90(0.00) | LDD0038 | [5] | |
|
IA-alkyne Probe Info |
 |
C438(0.00); C219(0.00); C132(0.00); C90(0.00) | LDD0036 | [5] | |
|
Lodoacetamide azide Probe Info |
 |
C438(0.00); C219(0.00); C132(0.00); C90(0.00) | LDD0037 | [5] | |
|
NAIA_4 Probe Info |
 |
C90(0.00); C474(0.00) | LDD2226 | [6] | |
|
KY-26 Probe Info |
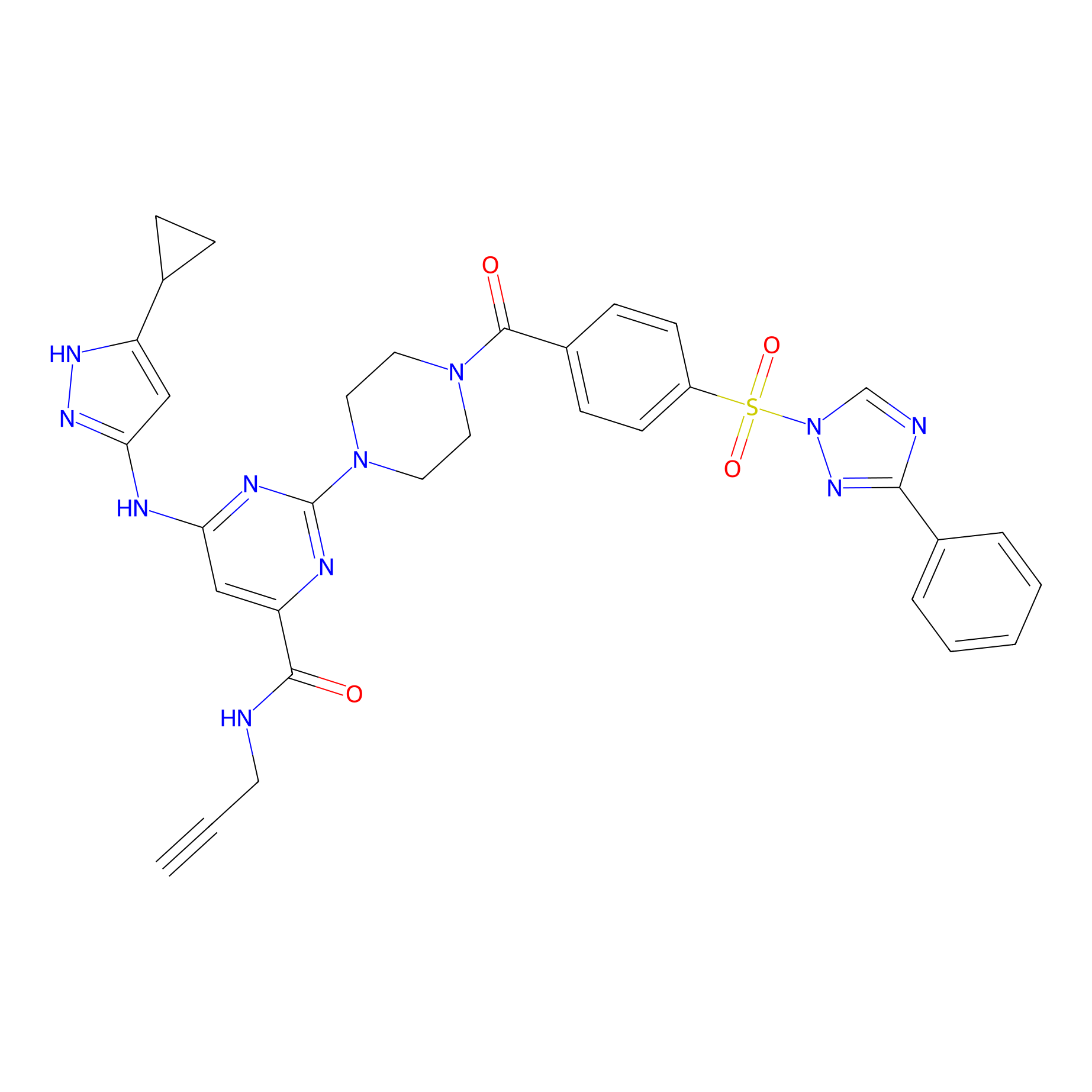 |
N.A. | LDD0301 | [7] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [8] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | DM93 | C474(3.29) | LDD0170 | [3] |
| LDCM0026 | 4SU-RNA+native RNA | DM93 | C219(4.97) | LDD0171 | [3] |
| LDCM0214 | AC1 | PaTu 8988t | C90(1.69) | LDD1093 | [9] |
| LDCM0215 | AC10 | PaTu 8988t | C144(0.62); C147(0.62); C90(1.16) | LDD1094 | [9] |
| LDCM0216 | AC100 | PaTu 8988t | C144(1.04); C147(1.04); C219(0.98); C90(0.99) | LDD1095 | [9] |
| LDCM0217 | AC101 | PaTu 8988t | C144(0.97); C147(0.97); C219(0.94); C90(1.06) | LDD1096 | [9] |
| LDCM0218 | AC102 | PaTu 8988t | C144(0.81); C147(0.81); C219(0.90); C90(1.22) | LDD1097 | [9] |
| LDCM0219 | AC103 | PaTu 8988t | C144(0.91); C147(0.91); C219(1.06); C90(1.30) | LDD1098 | [9] |
| LDCM0220 | AC104 | PaTu 8988t | C144(1.54); C147(1.54); C219(0.81); C90(1.04) | LDD1099 | [9] |
| LDCM0221 | AC105 | PaTu 8988t | C144(1.02); C147(1.02); C219(0.72); C90(0.97) | LDD1100 | [9] |
| LDCM0222 | AC106 | PaTu 8988t | C144(0.81); C147(0.81); C219(0.83); C90(1.14) | LDD1101 | [9] |
| LDCM0223 | AC107 | PaTu 8988t | C144(0.84); C147(0.84); C219(1.09); C90(0.91) | LDD1102 | [9] |
| LDCM0224 | AC108 | PaTu 8988t | C144(0.81); C147(0.81); C219(0.88); C90(1.12) | LDD1103 | [9] |
| LDCM0225 | AC109 | PaTu 8988t | C144(0.78); C147(0.78); C219(0.96); C90(1.20) | LDD1104 | [9] |
| LDCM0226 | AC11 | PaTu 8988t | C144(0.67); C147(0.67); C90(1.09) | LDD1105 | [9] |
| LDCM0227 | AC110 | PaTu 8988t | C144(0.72); C147(0.72); C219(1.16); C90(1.22) | LDD1106 | [9] |
| LDCM0228 | AC111 | PaTu 8988t | C144(0.74); C147(0.74); C219(0.55); C90(1.21) | LDD1107 | [9] |
| LDCM0229 | AC112 | PaTu 8988t | C144(0.79); C147(0.79); C219(0.75); C90(1.14) | LDD1108 | [9] |
| LDCM0230 | AC113 | PaTu 8988t | C144(1.35); C147(1.35); C90(1.00) | LDD1109 | [9] |
| LDCM0231 | AC114 | PaTu 8988t | C144(0.85); C147(0.85); C90(1.04) | LDD1110 | [9] |
| LDCM0232 | AC115 | PaTu 8988t | C144(1.10); C147(1.10); C90(1.17) | LDD1111 | [9] |
| LDCM0233 | AC116 | PaTu 8988t | C144(1.28); C147(1.28); C90(1.06) | LDD1112 | [9] |
| LDCM0234 | AC117 | PaTu 8988t | C144(1.27); C147(1.27); C90(1.07) | LDD1113 | [9] |
| LDCM0235 | AC118 | PaTu 8988t | C144(0.84); C147(0.84); C90(1.04) | LDD1114 | [9] |
| LDCM0236 | AC119 | PaTu 8988t | C144(1.11); C147(1.11); C90(1.03) | LDD1115 | [9] |
| LDCM0237 | AC12 | PaTu 8988t | C144(0.71); C147(0.71); C90(1.14) | LDD1116 | [9] |
| LDCM0238 | AC120 | PaTu 8988t | C144(1.33); C147(1.33); C90(0.92) | LDD1117 | [9] |
| LDCM0239 | AC121 | PaTu 8988t | C144(0.93); C147(0.93); C90(1.14) | LDD1118 | [9] |
| LDCM0240 | AC122 | PaTu 8988t | C144(1.02); C147(1.02); C90(0.94) | LDD1119 | [9] |
| LDCM0241 | AC123 | PaTu 8988t | C144(1.21); C147(1.21); C90(0.97) | LDD1120 | [9] |
| LDCM0242 | AC124 | PaTu 8988t | C144(0.97); C147(0.97); C90(1.05) | LDD1121 | [9] |
| LDCM0243 | AC125 | PaTu 8988t | C144(0.85); C147(0.85); C90(1.22) | LDD1122 | [9] |
| LDCM0244 | AC126 | PaTu 8988t | C144(1.01); C147(1.01); C90(0.98) | LDD1123 | [9] |
| LDCM0245 | AC127 | PaTu 8988t | C144(1.16); C147(1.16); C90(0.98) | LDD1124 | [9] |
| LDCM0246 | AC128 | PaTu 8988t | C144(0.93); C147(0.93); C90(1.03) | LDD1125 | [9] |
| LDCM0247 | AC129 | PaTu 8988t | C144(0.72); C147(0.72); C90(1.21) | LDD1126 | [9] |
| LDCM0249 | AC130 | PaTu 8988t | C144(0.80); C147(0.80); C90(1.00) | LDD1128 | [9] |
| LDCM0250 | AC131 | PaTu 8988t | C144(0.96); C147(0.96); C90(1.04) | LDD1129 | [9] |
| LDCM0251 | AC132 | PaTu 8988t | C144(0.79); C147(0.79); C90(1.22) | LDD1130 | [9] |
| LDCM0252 | AC133 | PaTu 8988t | C144(0.87); C147(0.87); C90(1.12) | LDD1131 | [9] |
| LDCM0253 | AC134 | PaTu 8988t | C144(0.90); C147(0.90); C90(1.06) | LDD1132 | [9] |
| LDCM0254 | AC135 | PaTu 8988t | C144(0.85); C147(0.85); C90(1.12) | LDD1133 | [9] |
| LDCM0255 | AC136 | PaTu 8988t | C144(0.91); C147(0.91); C90(1.26) | LDD1134 | [9] |
| LDCM0256 | AC137 | PaTu 8988t | C144(0.83); C147(0.83); C90(1.03) | LDD1135 | [9] |
| LDCM0257 | AC138 | PaTu 8988t | C144(0.84); C147(0.84); C90(1.11) | LDD1136 | [9] |
| LDCM0258 | AC139 | PaTu 8988t | C144(1.12); C147(1.12); C90(1.10) | LDD1137 | [9] |
| LDCM0259 | AC14 | PaTu 8988t | C144(0.67); C147(0.67); C90(0.81) | LDD1138 | [9] |
| LDCM0260 | AC140 | PaTu 8988t | C144(1.19); C147(1.19); C90(1.10) | LDD1139 | [9] |
| LDCM0261 | AC141 | PaTu 8988t | C144(0.94); C147(0.94); C90(1.22) | LDD1140 | [9] |
| LDCM0262 | AC142 | PaTu 8988t | C144(0.87); C147(0.87); C90(1.11) | LDD1141 | [9] |
| LDCM0263 | AC143 | PaTu 8988t | C144(0.74); C147(0.74); C219(0.89) | LDD1142 | [9] |
| LDCM0264 | AC144 | PaTu 8988t | C144(0.73); C147(0.73); C219(0.85) | LDD1143 | [9] |
| LDCM0265 | AC145 | PaTu 8988t | C144(0.80); C147(0.80); C219(1.07) | LDD1144 | [9] |
| LDCM0266 | AC146 | PaTu 8988t | C144(0.55); C147(0.55); C219(1.04) | LDD1145 | [9] |
| LDCM0267 | AC147 | PaTu 8988t | C144(0.71); C147(0.71); C219(1.00) | LDD1146 | [9] |
| LDCM0268 | AC148 | PaTu 8988t | C144(0.65); C147(0.65); C219(1.12) | LDD1147 | [9] |
| LDCM0269 | AC149 | PaTu 8988t | C144(0.67); C147(0.67); C219(1.19) | LDD1148 | [9] |
| LDCM0270 | AC15 | PaTu 8988t | C144(0.65); C147(0.65); C90(0.96) | LDD1149 | [9] |
| LDCM0271 | AC150 | PaTu 8988t | C144(0.53); C147(0.53); C219(1.23) | LDD1150 | [9] |
| LDCM0272 | AC151 | PaTu 8988t | C144(0.58); C147(0.58); C219(1.57) | LDD1151 | [9] |
| LDCM0273 | AC152 | PaTu 8988t | C144(0.64); C147(0.64); C219(1.07) | LDD1152 | [9] |
| LDCM0274 | AC153 | PaTu 8988t | C144(0.52); C147(0.52); C219(1.27) | LDD1153 | [9] |
| LDCM0621 | AC154 | PaTu 8988t | C144(0.58); C147(0.58); C219(1.51) | LDD2166 | [9] |
| LDCM0622 | AC155 | PaTu 8988t | C144(0.58); C147(0.58); C219(1.17) | LDD2167 | [9] |
| LDCM0623 | AC156 | PaTu 8988t | C144(0.62); C147(0.62); C219(1.17) | LDD2168 | [9] |
| LDCM0624 | AC157 | PaTu 8988t | C144(0.56); C147(0.56); C219(1.46) | LDD2169 | [9] |
| LDCM0276 | AC17 | PaTu 8988t | C219(0.82); C90(0.80) | LDD1155 | [9] |
| LDCM0277 | AC18 | PaTu 8988t | C219(0.86); C90(1.00) | LDD1156 | [9] |
| LDCM0278 | AC19 | PaTu 8988t | C219(1.00); C90(0.88) | LDD1157 | [9] |
| LDCM0279 | AC2 | PaTu 8988t | C90(1.41) | LDD1158 | [9] |
| LDCM0280 | AC20 | PaTu 8988t | C219(0.75); C90(1.00) | LDD1159 | [9] |
| LDCM0281 | AC21 | PaTu 8988t | C219(0.76); C90(0.86) | LDD1160 | [9] |
| LDCM0282 | AC22 | PaTu 8988t | C219(0.96); C90(1.03) | LDD1161 | [9] |
| LDCM0283 | AC23 | PaTu 8988t | C219(0.96); C90(1.27) | LDD1162 | [9] |
| LDCM0284 | AC24 | PaTu 8988t | C219(0.92); C90(1.03) | LDD1163 | [9] |
| LDCM0285 | AC25 | PaTu 8988t | C144(0.91); C147(0.91); C219(0.88); C90(0.98) | LDD1164 | [9] |
| LDCM0286 | AC26 | PaTu 8988t | C144(0.87); C147(0.87); C219(0.91); C90(1.04) | LDD1165 | [9] |
| LDCM0287 | AC27 | PaTu 8988t | C144(0.95); C147(0.95); C219(1.16); C90(1.06) | LDD1166 | [9] |
| LDCM0288 | AC28 | PaTu 8988t | C144(0.88); C147(0.88); C219(1.15); C90(1.07) | LDD1167 | [9] |
| LDCM0289 | AC29 | PaTu 8988t | C144(0.76); C147(0.76); C219(1.06); C90(1.02) | LDD1168 | [9] |
| LDCM0290 | AC3 | PaTu 8988t | C90(1.26) | LDD1169 | [9] |
| LDCM0291 | AC30 | PaTu 8988t | C144(1.26); C147(1.26); C219(1.07); C90(1.02) | LDD1170 | [9] |
| LDCM0292 | AC31 | PaTu 8988t | C144(0.87); C147(0.87); C219(0.92); C90(0.89) | LDD1171 | [9] |
| LDCM0293 | AC32 | PaTu 8988t | C144(0.79); C147(0.79); C219(0.91); C90(0.93) | LDD1172 | [9] |
| LDCM0294 | AC33 | PaTu 8988t | C144(0.75); C147(0.75); C219(1.23); C90(0.97) | LDD1173 | [9] |
| LDCM0295 | AC34 | PaTu 8988t | C144(0.68); C147(0.68); C219(0.91); C90(1.26) | LDD1174 | [9] |
| LDCM0296 | AC35 | PaTu 8988t | C144(1.37); C147(1.37); C90(1.06) | LDD1175 | [9] |
| LDCM0297 | AC36 | PaTu 8988t | C144(1.10); C147(1.10); C90(1.20) | LDD1176 | [9] |
| LDCM0298 | AC37 | PaTu 8988t | C144(0.83); C147(0.83); C90(1.23) | LDD1177 | [9] |
| LDCM0299 | AC38 | PaTu 8988t | C144(0.88); C147(0.88); C90(1.34) | LDD1178 | [9] |
| LDCM0300 | AC39 | PaTu 8988t | C144(1.09); C147(1.09); C90(1.24) | LDD1179 | [9] |
| LDCM0301 | AC4 | PaTu 8988t | C90(1.09) | LDD1180 | [9] |
| LDCM0302 | AC40 | PaTu 8988t | C144(0.75); C147(0.75); C90(1.46) | LDD1181 | [9] |
| LDCM0303 | AC41 | PaTu 8988t | C144(0.89); C147(0.89); C90(1.18) | LDD1182 | [9] |
| LDCM0304 | AC42 | PaTu 8988t | C144(0.71); C147(0.71); C90(1.30) | LDD1183 | [9] |
| LDCM0305 | AC43 | PaTu 8988t | C144(1.03); C147(1.03); C90(1.27) | LDD1184 | [9] |
| LDCM0306 | AC44 | PaTu 8988t | C144(0.71); C147(0.71); C90(1.36) | LDD1185 | [9] |
| LDCM0307 | AC45 | PaTu 8988t | C144(0.73); C147(0.73); C90(1.27) | LDD1186 | [9] |
| LDCM0308 | AC46 | PaTu 8988t | C219(1.08) | LDD1187 | [9] |
| LDCM0309 | AC47 | PaTu 8988t | C219(1.12) | LDD1188 | [9] |
| LDCM0310 | AC48 | PaTu 8988t | C219(1.19) | LDD1189 | [9] |
| LDCM0311 | AC49 | PaTu 8988t | C219(1.04) | LDD1190 | [9] |
| LDCM0312 | AC5 | PaTu 8988t | C90(1.55) | LDD1191 | [9] |
| LDCM0313 | AC50 | PaTu 8988t | C219(1.00) | LDD1192 | [9] |
| LDCM0314 | AC51 | PaTu 8988t | C219(0.94) | LDD1193 | [9] |
| LDCM0315 | AC52 | PaTu 8988t | C219(1.09) | LDD1194 | [9] |
| LDCM0316 | AC53 | PaTu 8988t | C219(1.11) | LDD1195 | [9] |
| LDCM0317 | AC54 | PaTu 8988t | C219(1.35) | LDD1196 | [9] |
| LDCM0318 | AC55 | PaTu 8988t | C219(0.97) | LDD1197 | [9] |
| LDCM0319 | AC56 | PaTu 8988t | C219(1.24) | LDD1198 | [9] |
| LDCM0320 | AC57 | PaTu 8988t | C90(1.25) | LDD1199 | [9] |
| LDCM0321 | AC58 | PaTu 8988t | C90(1.25) | LDD1200 | [9] |
| LDCM0322 | AC59 | PaTu 8988t | C90(1.13) | LDD1201 | [9] |
| LDCM0323 | AC6 | PaTu 8988t | C144(1.08); C147(1.08); C90(0.94) | LDD1202 | [9] |
| LDCM0324 | AC60 | PaTu 8988t | C90(1.36) | LDD1203 | [9] |
| LDCM0325 | AC61 | PaTu 8988t | C90(1.23) | LDD1204 | [9] |
| LDCM0326 | AC62 | PaTu 8988t | C90(1.17) | LDD1205 | [9] |
| LDCM0327 | AC63 | PaTu 8988t | C90(1.22) | LDD1206 | [9] |
| LDCM0328 | AC64 | PaTu 8988t | C90(1.15) | LDD1207 | [9] |
| LDCM0329 | AC65 | PaTu 8988t | C90(1.05) | LDD1208 | [9] |
| LDCM0330 | AC66 | PaTu 8988t | C90(1.20) | LDD1209 | [9] |
| LDCM0331 | AC67 | PaTu 8988t | C90(1.16) | LDD1210 | [9] |
| LDCM0332 | AC68 | PaTu 8988t | C144(1.00); C147(1.00); C219(1.00); C90(0.81) | LDD1211 | [9] |
| LDCM0333 | AC69 | PaTu 8988t | C144(0.89); C147(0.89); C219(0.89); C90(1.13) | LDD1212 | [9] |
| LDCM0334 | AC7 | PaTu 8988t | C144(1.23); C147(1.23); C90(0.98) | LDD1213 | [9] |
| LDCM0335 | AC70 | PaTu 8988t | C144(0.92); C147(0.92); C219(1.10); C90(0.98) | LDD1214 | [9] |
| LDCM0336 | AC71 | PaTu 8988t | C144(1.15); C147(1.15); C219(1.27); C90(1.16) | LDD1215 | [9] |
| LDCM0337 | AC72 | PaTu 8988t | C144(1.07); C147(1.07); C219(1.03); C90(0.95) | LDD1216 | [9] |
| LDCM0338 | AC73 | PaTu 8988t | C144(1.14); C147(1.14); C219(1.30); C90(0.84) | LDD1217 | [9] |
| LDCM0339 | AC74 | PaTu 8988t | C144(0.91); C147(0.91); C219(1.12); C90(1.12) | LDD1218 | [9] |
| LDCM0340 | AC75 | PaTu 8988t | C144(1.14); C147(1.14); C219(1.05); C90(1.12) | LDD1219 | [9] |
| LDCM0341 | AC76 | PaTu 8988t | C144(0.98); C147(0.98); C219(1.23); C90(1.00) | LDD1220 | [9] |
| LDCM0342 | AC77 | PaTu 8988t | C144(1.07); C147(1.07); C219(1.22); C90(1.00) | LDD1221 | [9] |
| LDCM0343 | AC78 | PaTu 8988t | C144(0.91); C147(0.91); C219(1.04); C90(1.01) | LDD1222 | [9] |
| LDCM0344 | AC79 | PaTu 8988t | C144(1.05); C147(1.05); C219(1.16); C90(1.16) | LDD1223 | [9] |
| LDCM0345 | AC8 | PaTu 8988t | C144(0.83); C147(0.83); C90(0.90) | LDD1224 | [9] |
| LDCM0346 | AC80 | PaTu 8988t | C144(0.84); C147(0.84); C219(1.14); C90(0.94) | LDD1225 | [9] |
| LDCM0347 | AC81 | PaTu 8988t | C144(0.96); C147(0.96); C219(1.46); C90(0.93) | LDD1226 | [9] |
| LDCM0348 | AC82 | PaTu 8988t | C144(0.83); C147(0.83); C219(1.27); C90(1.20) | LDD1227 | [9] |
| LDCM0349 | AC83 | PaTu 8988t | C219(0.97); C90(0.98) | LDD1228 | [9] |
| LDCM0350 | AC84 | PaTu 8988t | C219(1.11); C90(0.90) | LDD1229 | [9] |
| LDCM0351 | AC85 | PaTu 8988t | C219(0.89); C90(0.86) | LDD1230 | [9] |
| LDCM0352 | AC86 | PaTu 8988t | C219(0.89); C90(1.08) | LDD1231 | [9] |
| LDCM0353 | AC87 | PaTu 8988t | C219(1.04); C90(0.94) | LDD1232 | [9] |
| LDCM0354 | AC88 | PaTu 8988t | C219(1.12); C90(0.99) | LDD1233 | [9] |
| LDCM0355 | AC89 | PaTu 8988t | C219(0.99); C90(1.16) | LDD1234 | [9] |
| LDCM0357 | AC90 | PaTu 8988t | C219(1.45); C90(0.96) | LDD1236 | [9] |
| LDCM0358 | AC91 | PaTu 8988t | C219(0.94); C90(0.94) | LDD1237 | [9] |
| LDCM0359 | AC92 | PaTu 8988t | C219(0.81); C90(1.03) | LDD1238 | [9] |
| LDCM0360 | AC93 | PaTu 8988t | C219(0.95); C90(0.84) | LDD1239 | [9] |
| LDCM0361 | AC94 | PaTu 8988t | C219(1.14); C90(0.99) | LDD1240 | [9] |
| LDCM0362 | AC95 | PaTu 8988t | C219(0.83); C90(1.04) | LDD1241 | [9] |
| LDCM0363 | AC96 | PaTu 8988t | C219(1.18); C90(0.91) | LDD1242 | [9] |
| LDCM0364 | AC97 | PaTu 8988t | C219(0.92); C90(0.94) | LDD1243 | [9] |
| LDCM0365 | AC98 | PaTu 8988t | C144(0.75); C147(0.75); C219(0.98); C90(1.12) | LDD1244 | [9] |
| LDCM0366 | AC99 | PaTu 8988t | C144(0.73); C147(0.73); C219(0.75); C90(1.07) | LDD1245 | [9] |
| LDCM0248 | AKOS034007472 | PaTu 8988t | C144(0.59); C147(0.59); C90(0.90) | LDD1127 | [9] |
| LDCM0356 | AKOS034007680 | PaTu 8988t | C144(0.79); C147(0.79); C90(1.46) | LDD1235 | [9] |
| LDCM0275 | AKOS034007705 | PaTu 8988t | C144(0.64); C147(0.64); C90(1.28) | LDD1154 | [9] |
| LDCM0367 | CL1 | PaTu 8988t | C219(0.87) | LDD1246 | [9] |
| LDCM0368 | CL10 | PaTu 8988t | C219(1.06) | LDD1247 | [9] |
| LDCM0369 | CL100 | PaTu 8988t | C90(1.23) | LDD1248 | [9] |
| LDCM0370 | CL101 | PaTu 8988t | C144(0.95); C147(0.95); C90(1.33) | LDD1249 | [9] |
| LDCM0371 | CL102 | PaTu 8988t | C144(0.85); C147(0.85); C90(1.07) | LDD1250 | [9] |
| LDCM0372 | CL103 | PaTu 8988t | C144(0.75); C147(0.75); C90(1.26) | LDD1251 | [9] |
| LDCM0373 | CL104 | PaTu 8988t | C144(0.81); C147(0.81); C90(1.06) | LDD1252 | [9] |
| LDCM0374 | CL105 | PaTu 8988t | C219(0.69); C90(0.92) | LDD1253 | [9] |
| LDCM0375 | CL106 | PaTu 8988t | C219(0.88); C90(0.99) | LDD1254 | [9] |
| LDCM0376 | CL107 | PaTu 8988t | C219(0.95); C90(0.78) | LDD1255 | [9] |
| LDCM0377 | CL108 | PaTu 8988t | C219(1.00); C90(0.88) | LDD1256 | [9] |
| LDCM0378 | CL109 | PaTu 8988t | C219(0.96); C90(1.10) | LDD1257 | [9] |
| LDCM0379 | CL11 | PaTu 8988t | C219(1.07) | LDD1258 | [9] |
| LDCM0380 | CL110 | PaTu 8988t | C219(0.97); C90(1.08) | LDD1259 | [9] |
| LDCM0381 | CL111 | PaTu 8988t | C219(1.01); C90(1.40) | LDD1260 | [9] |
| LDCM0382 | CL112 | PaTu 8988t | C144(0.82); C147(0.82); C219(0.95); C90(0.91) | LDD1261 | [9] |
| LDCM0383 | CL113 | PaTu 8988t | C144(0.94); C147(0.94); C219(1.05); C90(0.94) | LDD1262 | [9] |
| LDCM0384 | CL114 | PaTu 8988t | C144(1.04); C147(1.04); C219(1.12); C90(0.99) | LDD1263 | [9] |
| LDCM0385 | CL115 | PaTu 8988t | C144(0.87); C147(0.87); C219(1.12); C90(0.92) | LDD1264 | [9] |
| LDCM0386 | CL116 | PaTu 8988t | C144(0.92); C147(0.92); C219(1.16); C90(0.98) | LDD1265 | [9] |
| LDCM0387 | CL117 | PaTu 8988t | C144(1.10); C147(1.10); C90(1.08) | LDD1266 | [9] |
| LDCM0388 | CL118 | PaTu 8988t | C144(0.64); C147(0.64); C90(1.02) | LDD1267 | [9] |
| LDCM0389 | CL119 | PaTu 8988t | C144(0.84); C147(0.84); C90(1.31) | LDD1268 | [9] |
| LDCM0390 | CL12 | PaTu 8988t | C219(0.82) | LDD1269 | [9] |
| LDCM0391 | CL120 | PaTu 8988t | C144(1.23); C147(1.23); C90(1.34) | LDD1270 | [9] |
| LDCM0392 | CL121 | PaTu 8988t | C219(1.00) | LDD1271 | [9] |
| LDCM0393 | CL122 | PaTu 8988t | C219(1.12) | LDD1272 | [9] |
| LDCM0394 | CL123 | PaTu 8988t | C219(1.11) | LDD1273 | [9] |
| LDCM0395 | CL124 | PaTu 8988t | C219(1.08) | LDD1274 | [9] |
| LDCM0396 | CL125 | PaTu 8988t | C90(1.19) | LDD1275 | [9] |
| LDCM0397 | CL126 | PaTu 8988t | C90(1.11) | LDD1276 | [9] |
| LDCM0398 | CL127 | PaTu 8988t | C90(1.24) | LDD1277 | [9] |
| LDCM0399 | CL128 | PaTu 8988t | C90(1.15) | LDD1278 | [9] |
| LDCM0400 | CL13 | PaTu 8988t | C219(1.24) | LDD1279 | [9] |
| LDCM0401 | CL14 | PaTu 8988t | C219(1.31) | LDD1280 | [9] |
| LDCM0402 | CL15 | PaTu 8988t | C219(0.89) | LDD1281 | [9] |
| LDCM0403 | CL16 | PaTu 8988t | C144(0.90); C147(0.90) | LDD1282 | [9] |
| LDCM0404 | CL17 | PaTu 8988t | C144(0.73); C147(0.73) | LDD1283 | [9] |
| LDCM0405 | CL18 | PaTu 8988t | C144(0.72); C147(0.72) | LDD1284 | [9] |
| LDCM0406 | CL19 | PaTu 8988t | C144(0.71); C147(0.71) | LDD1285 | [9] |
| LDCM0407 | CL2 | PaTu 8988t | C219(0.98) | LDD1286 | [9] |
| LDCM0408 | CL20 | PaTu 8988t | C144(0.74); C147(0.74) | LDD1287 | [9] |
| LDCM0409 | CL21 | PaTu 8988t | C144(0.77); C147(0.77) | LDD1288 | [9] |
| LDCM0410 | CL22 | PaTu 8988t | C144(0.71); C147(0.71) | LDD1289 | [9] |
| LDCM0411 | CL23 | PaTu 8988t | C144(0.77); C147(0.77) | LDD1290 | [9] |
| LDCM0412 | CL24 | PaTu 8988t | C144(0.77); C147(0.77) | LDD1291 | [9] |
| LDCM0413 | CL25 | PaTu 8988t | C144(0.84); C147(0.84) | LDD1292 | [9] |
| LDCM0414 | CL26 | PaTu 8988t | C144(0.78); C147(0.78) | LDD1293 | [9] |
| LDCM0415 | CL27 | PaTu 8988t | C144(0.76); C147(0.76) | LDD1294 | [9] |
| LDCM0416 | CL28 | PaTu 8988t | C144(0.83); C147(0.83) | LDD1295 | [9] |
| LDCM0417 | CL29 | PaTu 8988t | C144(0.86); C147(0.86) | LDD1296 | [9] |
| LDCM0418 | CL3 | PaTu 8988t | C219(0.94) | LDD1297 | [9] |
| LDCM0419 | CL30 | PaTu 8988t | C144(0.71); C147(0.71) | LDD1298 | [9] |
| LDCM0420 | CL31 | PaTu 8988t | C144(0.88); C147(0.88); C219(1.08) | LDD1299 | [9] |
| LDCM0421 | CL32 | PaTu 8988t | C144(1.07); C147(1.07); C219(1.11) | LDD1300 | [9] |
| LDCM0422 | CL33 | PaTu 8988t | C144(0.86); C147(0.86); C219(1.27) | LDD1301 | [9] |
| LDCM0423 | CL34 | PaTu 8988t | C144(0.77); C147(0.77); C219(1.08) | LDD1302 | [9] |
| LDCM0424 | CL35 | PaTu 8988t | C144(0.78); C147(0.78); C219(1.06) | LDD1303 | [9] |
| LDCM0425 | CL36 | PaTu 8988t | C144(0.80); C147(0.80); C219(0.55) | LDD1304 | [9] |
| LDCM0426 | CL37 | PaTu 8988t | C144(0.86); C147(0.86); C219(1.01) | LDD1305 | [9] |
| LDCM0428 | CL39 | PaTu 8988t | C144(0.87); C147(0.87); C219(1.20) | LDD1307 | [9] |
| LDCM0429 | CL4 | PaTu 8988t | C219(0.74) | LDD1308 | [9] |
| LDCM0430 | CL40 | PaTu 8988t | C144(0.80); C147(0.80); C219(1.04) | LDD1309 | [9] |
| LDCM0431 | CL41 | PaTu 8988t | C144(0.77); C147(0.77); C219(1.24) | LDD1310 | [9] |
| LDCM0432 | CL42 | PaTu 8988t | C144(0.76); C147(0.76); C219(1.18) | LDD1311 | [9] |
| LDCM0433 | CL43 | PaTu 8988t | C144(0.89); C147(0.89); C219(1.24) | LDD1312 | [9] |
| LDCM0434 | CL44 | PaTu 8988t | C144(0.72); C147(0.72); C219(1.39) | LDD1313 | [9] |
| LDCM0435 | CL45 | PaTu 8988t | C144(0.71); C147(0.71); C219(1.31) | LDD1314 | [9] |
| LDCM0440 | CL5 | PaTu 8988t | C219(1.14) | LDD1319 | [9] |
| LDCM0451 | CL6 | PaTu 8988t | C219(1.11) | LDD1330 | [9] |
| LDCM0453 | CL61 | PaTu 8988t | C219(1.00); C90(0.97) | LDD1332 | [9] |
| LDCM0454 | CL62 | PaTu 8988t | C219(0.86); C90(1.27) | LDD1333 | [9] |
| LDCM0455 | CL63 | PaTu 8988t | C219(0.78); C90(1.41) | LDD1334 | [9] |
| LDCM0456 | CL64 | PaTu 8988t | C219(0.95); C90(1.20) | LDD1335 | [9] |
| LDCM0457 | CL65 | PaTu 8988t | C219(1.14); C90(1.53) | LDD1336 | [9] |
| LDCM0458 | CL66 | PaTu 8988t | C219(0.88); C90(1.65) | LDD1337 | [9] |
| LDCM0459 | CL67 | PaTu 8988t | C219(1.27); C90(1.24) | LDD1338 | [9] |
| LDCM0460 | CL68 | PaTu 8988t | C219(1.15); C90(1.36) | LDD1339 | [9] |
| LDCM0461 | CL69 | PaTu 8988t | C219(1.25); C90(1.91) | LDD1340 | [9] |
| LDCM0462 | CL7 | PaTu 8988t | C219(1.42) | LDD1341 | [9] |
| LDCM0463 | CL70 | PaTu 8988t | C219(1.17); C90(1.38) | LDD1342 | [9] |
| LDCM0464 | CL71 | PaTu 8988t | C219(0.81); C90(1.22) | LDD1343 | [9] |
| LDCM0465 | CL72 | PaTu 8988t | C219(0.70); C90(1.32) | LDD1344 | [9] |
| LDCM0466 | CL73 | PaTu 8988t | C219(1.09); C90(1.91) | LDD1345 | [9] |
| LDCM0467 | CL74 | PaTu 8988t | C219(0.83); C90(1.46) | LDD1346 | [9] |
| LDCM0469 | CL76 | PaTu 8988t | C144(0.66); C147(0.66); C219(0.99) | LDD1348 | [9] |
| LDCM0470 | CL77 | PaTu 8988t | C144(0.90); C147(0.90); C219(1.19) | LDD1349 | [9] |
| LDCM0471 | CL78 | PaTu 8988t | C144(0.87); C147(0.87); C219(1.42) | LDD1350 | [9] |
| LDCM0472 | CL79 | PaTu 8988t | C144(0.80); C147(0.80); C219(1.54) | LDD1351 | [9] |
| LDCM0473 | CL8 | PaTu 8988t | C219(1.10) | LDD1352 | [9] |
| LDCM0474 | CL80 | PaTu 8988t | C144(0.61); C147(0.61); C219(1.23) | LDD1353 | [9] |
| LDCM0475 | CL81 | PaTu 8988t | C144(0.69); C147(0.69); C219(1.40) | LDD1354 | [9] |
| LDCM0476 | CL82 | PaTu 8988t | C144(0.67); C147(0.67); C219(1.10) | LDD1355 | [9] |
| LDCM0477 | CL83 | PaTu 8988t | C144(0.90); C147(0.90); C219(1.29) | LDD1356 | [9] |
| LDCM0478 | CL84 | PaTu 8988t | C144(0.70); C147(0.70); C219(1.05) | LDD1357 | [9] |
| LDCM0479 | CL85 | PaTu 8988t | C144(0.85); C147(0.85); C219(1.48) | LDD1358 | [9] |
| LDCM0480 | CL86 | PaTu 8988t | C144(0.70); C147(0.70); C219(1.67) | LDD1359 | [9] |
| LDCM0481 | CL87 | PaTu 8988t | C144(0.65); C147(0.65); C219(1.54) | LDD1360 | [9] |
| LDCM0482 | CL88 | PaTu 8988t | C144(0.67); C147(0.67); C219(1.35) | LDD1361 | [9] |
| LDCM0483 | CL89 | PaTu 8988t | C144(0.68); C147(0.68); C219(1.23) | LDD1362 | [9] |
| LDCM0484 | CL9 | PaTu 8988t | C219(1.08) | LDD1363 | [9] |
| LDCM0485 | CL90 | PaTu 8988t | C144(0.76); C147(0.76); C219(1.40) | LDD1364 | [9] |
| LDCM0486 | CL91 | PaTu 8988t | C90(1.44) | LDD1365 | [9] |
| LDCM0487 | CL92 | PaTu 8988t | C90(1.41) | LDD1366 | [9] |
| LDCM0488 | CL93 | PaTu 8988t | C90(1.17) | LDD1367 | [9] |
| LDCM0489 | CL94 | PaTu 8988t | C90(1.42) | LDD1368 | [9] |
| LDCM0490 | CL95 | PaTu 8988t | C90(1.51) | LDD1369 | [9] |
| LDCM0491 | CL96 | PaTu 8988t | C90(1.48) | LDD1370 | [9] |
| LDCM0492 | CL97 | PaTu 8988t | C90(1.24) | LDD1371 | [9] |
| LDCM0493 | CL98 | PaTu 8988t | C90(1.57) | LDD1372 | [9] |
| LDCM0494 | CL99 | PaTu 8988t | C90(1.24) | LDD1373 | [9] |
| LDCM0495 | E2913 | HEK-293T | C90(1.01) | LDD1698 | [10] |
| LDCM0625 | F8 | Ramos | C90(1.23); C219(0.73); C366(1.47) | LDD2187 | [11] |
| LDCM0572 | Fragment10 | Ramos | C90(0.87); C219(0.68); C366(0.67) | LDD2189 | [11] |
| LDCM0573 | Fragment11 | Ramos | C90(0.17); C219(20.00); C366(1.91) | LDD2190 | [11] |
| LDCM0574 | Fragment12 | Ramos | C90(0.90); C219(0.65); C366(0.51) | LDD2191 | [11] |
| LDCM0575 | Fragment13 | Ramos | C90(1.01); C219(0.86); C366(0.99) | LDD2192 | [11] |
| LDCM0576 | Fragment14 | Ramos | C90(0.69); C219(1.77) | LDD2193 | [11] |
| LDCM0579 | Fragment20 | Ramos | C90(0.60); C219(0.70); C366(0.56) | LDD2194 | [11] |
| LDCM0580 | Fragment21 | Ramos | C90(0.94); C219(0.55) | LDD2195 | [11] |
| LDCM0582 | Fragment23 | Ramos | C90(1.01); C219(0.73) | LDD2196 | [11] |
| LDCM0578 | Fragment27 | Ramos | C90(0.83); C219(0.66); C366(0.95) | LDD2197 | [11] |
| LDCM0586 | Fragment28 | Ramos | C90(0.80); C219(0.50); C366(0.63) | LDD2198 | [11] |
| LDCM0588 | Fragment30 | Ramos | C90(0.95); C219(0.95); C366(0.77) | LDD2199 | [11] |
| LDCM0589 | Fragment31 | Ramos | C90(0.87); C219(0.90); C366(0.75) | LDD2200 | [11] |
| LDCM0590 | Fragment32 | Ramos | C90(1.53); C219(0.50); C366(0.61) | LDD2201 | [11] |
| LDCM0468 | Fragment33 | PaTu 8988t | C219(0.91); C90(1.68) | LDD1347 | [9] |
| LDCM0596 | Fragment38 | Ramos | C90(0.59); C219(0.44); C366(0.45) | LDD2203 | [11] |
| LDCM0566 | Fragment4 | Ramos | C90(1.42); C219(1.09) | LDD2184 | [11] |
| LDCM0427 | Fragment51 | PaTu 8988t | C144(0.76); C147(0.76); C219(1.09) | LDD1306 | [9] |
| LDCM0610 | Fragment52 | Ramos | C90(0.82); C219(0.58); C366(1.06) | LDD2204 | [11] |
| LDCM0614 | Fragment56 | Ramos | C90(0.89); C219(1.80) | LDD2205 | [11] |
| LDCM0569 | Fragment7 | Jurkat | _(6.74) | LDD1485 | [2] |
| LDCM0571 | Fragment9 | Ramos | C90(0.66); C219(0.84); C366(0.63) | LDD2188 | [11] |
| LDCM0022 | KB02 | Ramos | C90(1.10); C219(0.91); C366(0.65) | LDD2182 | [11] |
| LDCM0023 | KB03 | Jurkat | C991(6.96) | LDD0209 | [4] |
| LDCM0024 | KB05 | MKN-1 | C219(4.71); C474(0.69); C366(1.25) | LDD3331 | [12] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C991(0.56) | LDD2140 | [1] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| GTPase HRas (HRAS) | Ras family | P01112 | |||
Other
The Drug(s) Related To This Target
Approved
Phase 3
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Buparlisib | Small molecular drug | D01QSO | |||
Phase 2
Phase 1
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Bgb-10188 | Small molecular drug | DA02DE | |||
| Hmpl-689 | Small molecular drug | DDE3Z4 | |||
| Amg 319 | . | D03FOP | |||
| Gs-9820 | . | D03IJO | |||
| Gs-9901 | . | D0U5PT | |||
| Me-401 | . | D08WLY | |||
Preclinical
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Xl-499 | . | D08TAB | |||
Investigative
Patented
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Pyrrolo[2,3-d]Pyrimidine Derivative 28 | Small molecular drug | D00GTY | |||
Discontinued
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Wortmannin | Small molecular drug | D0P1JI | |||
References
