Details of the Target
General Information of Target
| Target ID | LDTP09764 | |||||
|---|---|---|---|---|---|---|
| Target Name | Acid sphingomyelinase-like phosphodiesterase 3b (SMPDL3B) | |||||
| Gene Name | SMPDL3B | |||||
| Gene ID | 27293 | |||||
| Synonyms |
ASML3B; ASMLPD; Acid sphingomyelinase-like phosphodiesterase 3b; ASM-like phosphodiesterase 3b; EC 3.1.4.- |
|||||
| 3D Structure | ||||||
| Sequence |
MALVRALVCCLLTAWHCRSGLGLPVAPAGGRNPPPAIGQFWHVTDLHLDPTYHITDDHTK
VCASSKGANASNPGPFGDVLCDSPYQLILSAFDFIKNSGQEASFMIWTGDSPPHVPVPEL STDTVINVITNMTTTIQSLFPNLQVFPALGNHDYWPQDQLPVVTSKVYNAVANLWKPWLD EEAISTLRKGGFYSQKVTTNPNLRIISLNTNLYYGPNIMTLNKTDPANQFEWLESTLNNS QQNKEKVYIIAHVPVGYLPSSQNITAMREYYNEKLIDIFQKYSDVIAGQFYGHTHRDSIM VLSDKKGSPVNSLFVAPAVTPVKSVLEKQTNNPGIRLFQYDPRDYKLLDMLQYYLNLTEA NLKGESIWKLEYILTQTYDIEDLQPESLYGLAKQFTILDSKQFIKYYNYFFVSYDSSVTC DKTCKAFQICAIMNLDNISYADCLKQLYIKHNY |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Acid sphingomyelinase family
|
|||||
| Subcellular location |
Secreted
|
|||||
| Function |
Lipid-modulating phosphodiesterase. Active on the surface of macrophages and dendritic cells and strongly influences macrophage lipid composition and membrane fluidity. Acts as a negative regulator of Toll-like receptor signaling. Has in vitro phosphodiesterase activity, but the physiological substrate is unknown. Lacks activity with phosphocholine-containing lipids, but can cleave CDP-choline, and can release phosphate from ATP and ADP (in vitro).
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DBIA Probe Info |
 |
C177(1.74) | LDD0532 | [1] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C170 Probe Info |
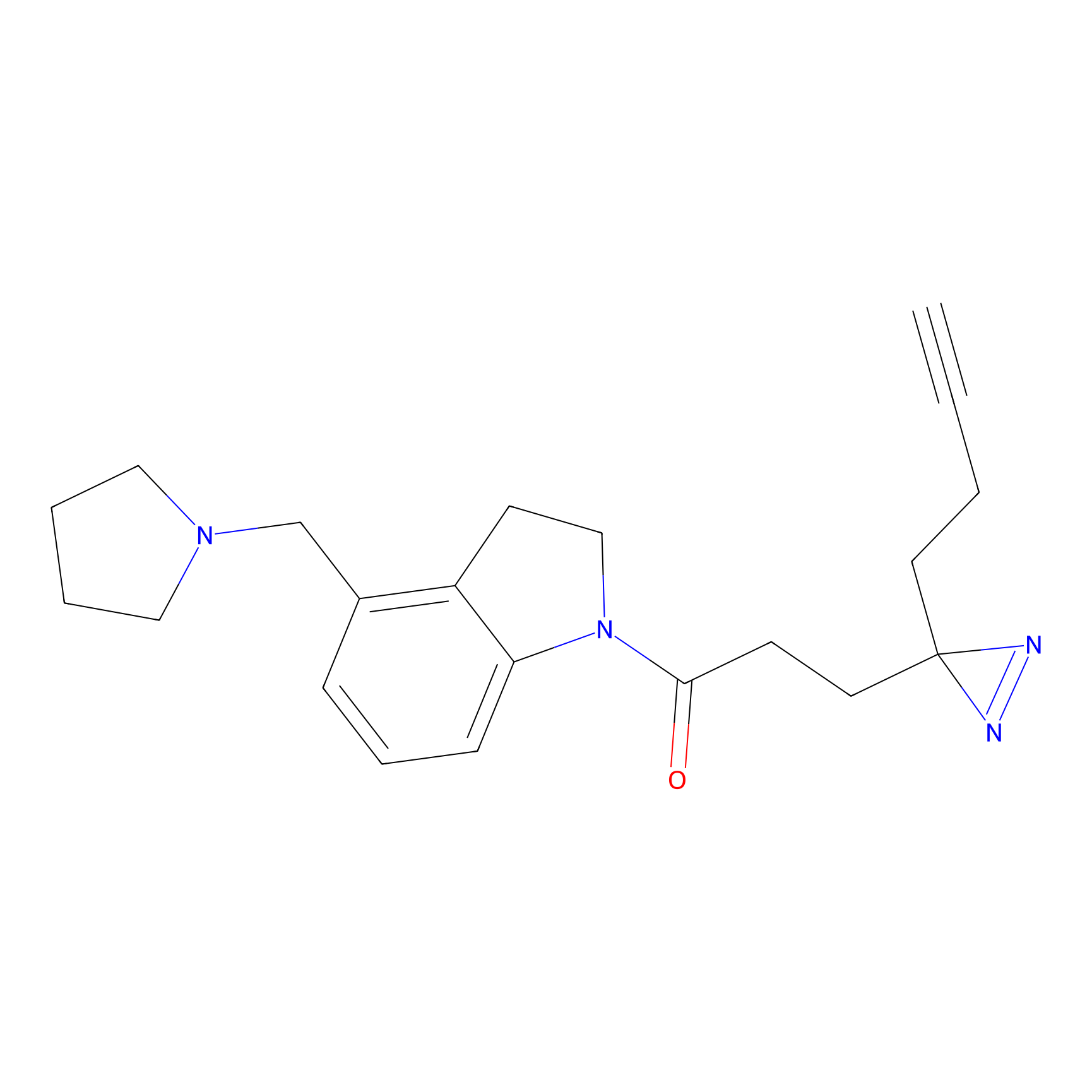 |
14.93 | LDD1850 | [2] | |
|
C171 Probe Info |
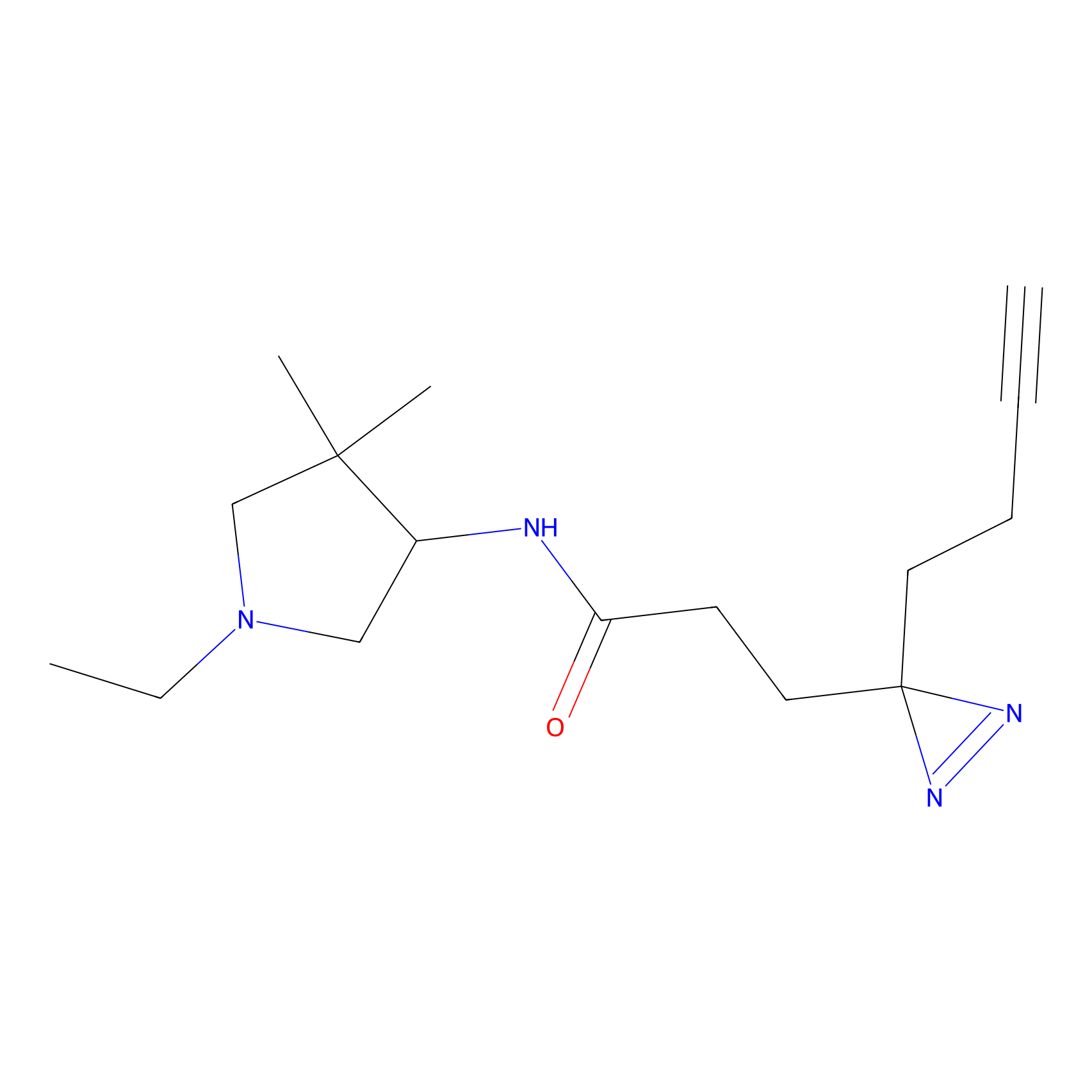 |
6.28 | LDD1851 | [2] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HEK-293T | C177(0.89) | LDD1507 | [3] |
| LDCM0215 | AC10 | HCT 116 | C177(1.74) | LDD0532 | [1] |
| LDCM0216 | AC100 | HCT 116 | C177(0.86) | LDD0533 | [1] |
| LDCM0217 | AC101 | HCT 116 | C177(1.09) | LDD0534 | [1] |
| LDCM0218 | AC102 | HCT 116 | C177(0.93) | LDD0535 | [1] |
| LDCM0219 | AC103 | HCT 116 | C177(0.98) | LDD0536 | [1] |
| LDCM0220 | AC104 | HCT 116 | C177(0.80) | LDD0537 | [1] |
| LDCM0221 | AC105 | HCT 116 | C177(1.08) | LDD0538 | [1] |
| LDCM0222 | AC106 | HCT 116 | C177(0.90) | LDD0539 | [1] |
| LDCM0223 | AC107 | HCT 116 | C177(1.06) | LDD0540 | [1] |
| LDCM0224 | AC108 | HCT 116 | C177(1.07) | LDD0541 | [1] |
| LDCM0225 | AC109 | HCT 116 | C177(0.88) | LDD0542 | [1] |
| LDCM0226 | AC11 | HCT 116 | C177(1.11) | LDD0543 | [1] |
| LDCM0227 | AC110 | HCT 116 | C177(0.89) | LDD0544 | [1] |
| LDCM0228 | AC111 | HCT 116 | C177(0.98) | LDD0545 | [1] |
| LDCM0229 | AC112 | HCT 116 | C177(1.07) | LDD0546 | [1] |
| LDCM0237 | AC12 | HCT 116 | C177(0.97) | LDD0554 | [1] |
| LDCM0259 | AC14 | HCT 116 | C177(1.34) | LDD0576 | [1] |
| LDCM0270 | AC15 | HCT 116 | C177(1.56) | LDD0587 | [1] |
| LDCM0276 | AC17 | HEK-293T | C177(0.98) | LDD1515 | [3] |
| LDCM0280 | AC20 | HEK-293T | C177(1.04) | LDD1519 | [3] |
| LDCM0282 | AC22 | HEK-293T | C177(1.09) | LDD1521 | [3] |
| LDCM0284 | AC24 | HEK-293T | C177(1.10) | LDD1523 | [3] |
| LDCM0285 | AC25 | HEK-293T | C177(0.89) | LDD1524 | [3] |
| LDCM0288 | AC28 | HEK-293T | C177(0.94) | LDD1527 | [3] |
| LDCM0291 | AC30 | HEK-293T | C177(0.94) | LDD1530 | [3] |
| LDCM0293 | AC32 | HEK-293T | C177(1.08) | LDD1532 | [3] |
| LDCM0294 | AC33 | HEK-293T | C177(1.11) | LDD1533 | [3] |
| LDCM0297 | AC36 | HEK-293T | C177(1.17) | LDD1536 | [3] |
| LDCM0299 | AC38 | HEK-293T | C177(1.04) | LDD1538 | [3] |
| LDCM0301 | AC4 | HEK-293T | C177(1.02) | LDD1540 | [3] |
| LDCM0302 | AC40 | HEK-293T | C177(1.00) | LDD1541 | [3] |
| LDCM0303 | AC41 | HEK-293T | C177(1.10) | LDD1542 | [3] |
| LDCM0306 | AC44 | HEK-293T | C177(1.21) | LDD1545 | [3] |
| LDCM0308 | AC46 | HEK-293T | C177(1.00) | LDD1547 | [3] |
| LDCM0310 | AC48 | HEK-293T | C177(1.12) | LDD1549 | [3] |
| LDCM0311 | AC49 | HEK-293T | C177(1.00) | LDD1550 | [3] |
| LDCM0315 | AC52 | HEK-293T | C177(1.20) | LDD1554 | [3] |
| LDCM0317 | AC54 | HEK-293T | C177(1.08) | LDD1556 | [3] |
| LDCM0319 | AC56 | HEK-293T | C177(1.13) | LDD1558 | [3] |
| LDCM0320 | AC57 | HEK-293T | C177(0.95) | LDD1559 | [3] |
| LDCM0323 | AC6 | HCT 116 | C177(1.27) | LDD0640 | [1] |
| LDCM0324 | AC60 | HEK-293T | C177(1.08) | LDD1563 | [3] |
| LDCM0326 | AC62 | HEK-293T | C177(1.04) | LDD1565 | [3] |
| LDCM0328 | AC64 | HEK-293T | C177(0.98) | LDD1567 | [3] |
| LDCM0334 | AC7 | HCT 116 | C177(1.00) | LDD0651 | [1] |
| LDCM0345 | AC8 | HCT 116 | C177(1.00) | LDD0662 | [1] |
| LDCM0365 | AC98 | HCT 116 | C177(1.14) | LDD0682 | [1] |
| LDCM0366 | AC99 | HCT 116 | C177(0.79) | LDD0683 | [1] |
| LDCM0248 | AKOS034007472 | HCT 116 | C177(1.13) | LDD0565 | [1] |
| LDCM0356 | AKOS034007680 | HCT 116 | C177(1.38) | LDD0673 | [1] |
| LDCM0275 | AKOS034007705 | HCT 116 | C177(2.21) | LDD0592 | [1] |
| LDCM0367 | CL1 | HEK-293T | C177(0.95) | LDD1571 | [3] |
| LDCM0368 | CL10 | HEK-293T | C177(0.89) | LDD1572 | [3] |
| LDCM0370 | CL101 | HCT 116 | C177(1.10) | LDD0687 | [1] |
| LDCM0371 | CL102 | HCT 116 | C177(0.96) | LDD0688 | [1] |
| LDCM0372 | CL103 | HCT 116 | C177(0.80) | LDD0689 | [1] |
| LDCM0373 | CL104 | HCT 116 | C177(1.15) | LDD0690 | [1] |
| LDCM0374 | CL105 | HEK-293T | C177(1.06) | LDD1578 | [3] |
| LDCM0378 | CL109 | HEK-293T | C177(0.97) | LDD1582 | [3] |
| LDCM0383 | CL113 | HEK-293T | C177(1.03) | LDD1587 | [3] |
| LDCM0387 | CL117 | HEK-293T | C177(1.07) | LDD1591 | [3] |
| LDCM0390 | CL12 | HEK-293T | C177(0.93) | LDD1594 | [3] |
| LDCM0392 | CL121 | HEK-293T | C177(1.04) | LDD1596 | [3] |
| LDCM0396 | CL125 | HEK-293T | C177(1.02) | LDD1600 | [3] |
| LDCM0400 | CL13 | HEK-293T | C177(0.91) | LDD1604 | [3] |
| LDCM0404 | CL17 | HEK-293T | C177(0.95) | LDD1608 | [3] |
| LDCM0408 | CL20 | HEK-293T | C177(1.10) | LDD1612 | [3] |
| LDCM0410 | CL22 | HEK-293T | C177(0.95) | LDD1614 | [3] |
| LDCM0412 | CL24 | HEK-293T | C177(1.04) | LDD1616 | [3] |
| LDCM0413 | CL25 | HEK-293T | C177(1.09) | LDD1617 | [3] |
| LDCM0417 | CL29 | HEK-293T | C177(1.01) | LDD1621 | [3] |
| LDCM0420 | CL31 | HCT 116 | C177(0.81) | LDD0737 | [1] |
| LDCM0421 | CL32 | HCT 116 | C177(0.96) | LDD0738 | [1] |
| LDCM0422 | CL33 | HCT 116 | C177(0.71) | LDD0739 | [1] |
| LDCM0423 | CL34 | HCT 116 | C177(1.00) | LDD0740 | [1] |
| LDCM0424 | CL35 | HCT 116 | C177(0.88) | LDD0741 | [1] |
| LDCM0425 | CL36 | HCT 116 | C177(0.80) | LDD0742 | [1] |
| LDCM0426 | CL37 | HCT 116 | C177(0.98) | LDD0743 | [1] |
| LDCM0428 | CL39 | HCT 116 | C177(1.17) | LDD0745 | [1] |
| LDCM0430 | CL40 | HCT 116 | C177(0.94) | LDD0747 | [1] |
| LDCM0431 | CL41 | HCT 116 | C177(1.25) | LDD0748 | [1] |
| LDCM0432 | CL42 | HCT 116 | C177(1.53) | LDD0749 | [1] |
| LDCM0433 | CL43 | HCT 116 | C177(1.01) | LDD0750 | [1] |
| LDCM0434 | CL44 | HCT 116 | C177(0.87) | LDD0751 | [1] |
| LDCM0435 | CL45 | HCT 116 | C177(1.02) | LDD0752 | [1] |
| LDCM0436 | CL46 | HCT 116 | C177(0.99) | LDD0753 | [1] |
| LDCM0437 | CL47 | HCT 116 | C177(0.89) | LDD0754 | [1] |
| LDCM0438 | CL48 | HCT 116 | C177(0.74) | LDD0755 | [1] |
| LDCM0439 | CL49 | HCT 116 | C177(0.61) | LDD0756 | [1] |
| LDCM0440 | CL5 | HEK-293T | C177(0.95) | LDD1644 | [3] |
| LDCM0441 | CL50 | HCT 116 | C177(0.94) | LDD0758 | [1] |
| LDCM0442 | CL51 | HCT 116 | C177(0.85) | LDD0759 | [1] |
| LDCM0443 | CL52 | HCT 116 | C177(0.78) | LDD0760 | [1] |
| LDCM0444 | CL53 | HCT 116 | C177(0.77) | LDD0761 | [1] |
| LDCM0445 | CL54 | HCT 116 | C177(0.81) | LDD0762 | [1] |
| LDCM0446 | CL55 | HCT 116 | C177(0.78) | LDD0763 | [1] |
| LDCM0447 | CL56 | HCT 116 | C177(1.06) | LDD0764 | [1] |
| LDCM0448 | CL57 | HCT 116 | C177(0.88) | LDD0765 | [1] |
| LDCM0449 | CL58 | HCT 116 | C177(0.93) | LDD0766 | [1] |
| LDCM0450 | CL59 | HCT 116 | C177(0.93) | LDD0767 | [1] |
| LDCM0452 | CL60 | HCT 116 | C177(0.94) | LDD0769 | [1] |
| LDCM0453 | CL61 | HCT 116 | C177(1.26) | LDD0770 | [1] |
| LDCM0454 | CL62 | HCT 116 | C177(1.30) | LDD0771 | [1] |
| LDCM0455 | CL63 | HCT 116 | C177(1.14) | LDD0772 | [1] |
| LDCM0456 | CL64 | HCT 116 | C177(1.24) | LDD0773 | [1] |
| LDCM0457 | CL65 | HCT 116 | C177(1.24) | LDD0774 | [1] |
| LDCM0458 | CL66 | HCT 116 | C177(1.88) | LDD0775 | [1] |
| LDCM0459 | CL67 | HCT 116 | C177(1.57) | LDD0776 | [1] |
| LDCM0460 | CL68 | HCT 116 | C177(1.62) | LDD0777 | [1] |
| LDCM0461 | CL69 | HCT 116 | C177(1.10) | LDD0778 | [1] |
| LDCM0463 | CL70 | HCT 116 | C177(1.23) | LDD0780 | [1] |
| LDCM0464 | CL71 | HCT 116 | C177(1.39) | LDD0781 | [1] |
| LDCM0465 | CL72 | HCT 116 | C177(1.49) | LDD0782 | [1] |
| LDCM0466 | CL73 | HCT 116 | C177(1.33) | LDD0783 | [1] |
| LDCM0467 | CL74 | HCT 116 | C177(1.57) | LDD0784 | [1] |
| LDCM0470 | CL77 | HEK-293T | C177(0.90) | LDD1673 | [3] |
| LDCM0473 | CL8 | HEK-293T | C177(0.86) | LDD1676 | [3] |
| LDCM0474 | CL80 | HEK-293T | C177(1.13) | LDD1677 | [3] |
| LDCM0476 | CL82 | HEK-293T | C177(0.98) | LDD1679 | [3] |
| LDCM0478 | CL84 | HEK-293T | C177(0.96) | LDD1681 | [3] |
| LDCM0479 | CL85 | HEK-293T | C177(1.09) | LDD1682 | [3] |
| LDCM0483 | CL89 | HEK-293T | C177(1.13) | LDD1686 | [3] |
| LDCM0487 | CL92 | HEK-293T | C177(1.19) | LDD1690 | [3] |
| LDCM0489 | CL94 | HEK-293T | C177(1.05) | LDD1692 | [3] |
| LDCM0491 | CL96 | HEK-293T | C177(0.98) | LDD1694 | [3] |
| LDCM0492 | CL97 | HEK-293T | C177(0.92) | LDD1695 | [3] |
| LDCM0468 | Fragment33 | HCT 116 | C177(1.44) | LDD0785 | [1] |
| LDCM0427 | Fragment51 | HCT 116 | C177(1.20) | LDD0744 | [1] |
| LDCM0022 | KB02 | 22RV1 | C177(1.32) | LDD2243 | [4] |
| LDCM0023 | KB03 | 22RV1 | C177(1.57) | LDD2660 | [4] |
| LDCM0024 | KB05 | OVCAR-3 | C177(1.10) | LDD3365 | [4] |
References
