Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TH211 Probe Info |
 |
Y137(20.00); Y32(9.09); Y106(7.65); Y38(7.01) | LDD0260 | [1] | |
|
STPyne Probe Info |
 |
K101(2.70); K140(5.79); K20(3.52); K39(0.89) | LDD0277 | [2] | |
|
5E-2FA Probe Info |
 |
H22(0.00); H132(0.00) | LDD2235 | [3] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [4] | |
|
m-APA Probe Info |
 |
H22(0.00); H132(0.00) | LDD2231 | [3] | |
|
N1 Probe Info |
 |
N.A. | LDD0245 | [5] | |
|
NHS Probe Info |
 |
K140(0.00); K80(0.00) | LDD0010 | [6] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [7] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [8] | |
|
Acrolein Probe Info |
 |
H22(0.00); H132(0.00) | LDD0217 | [9] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C003 Probe Info |
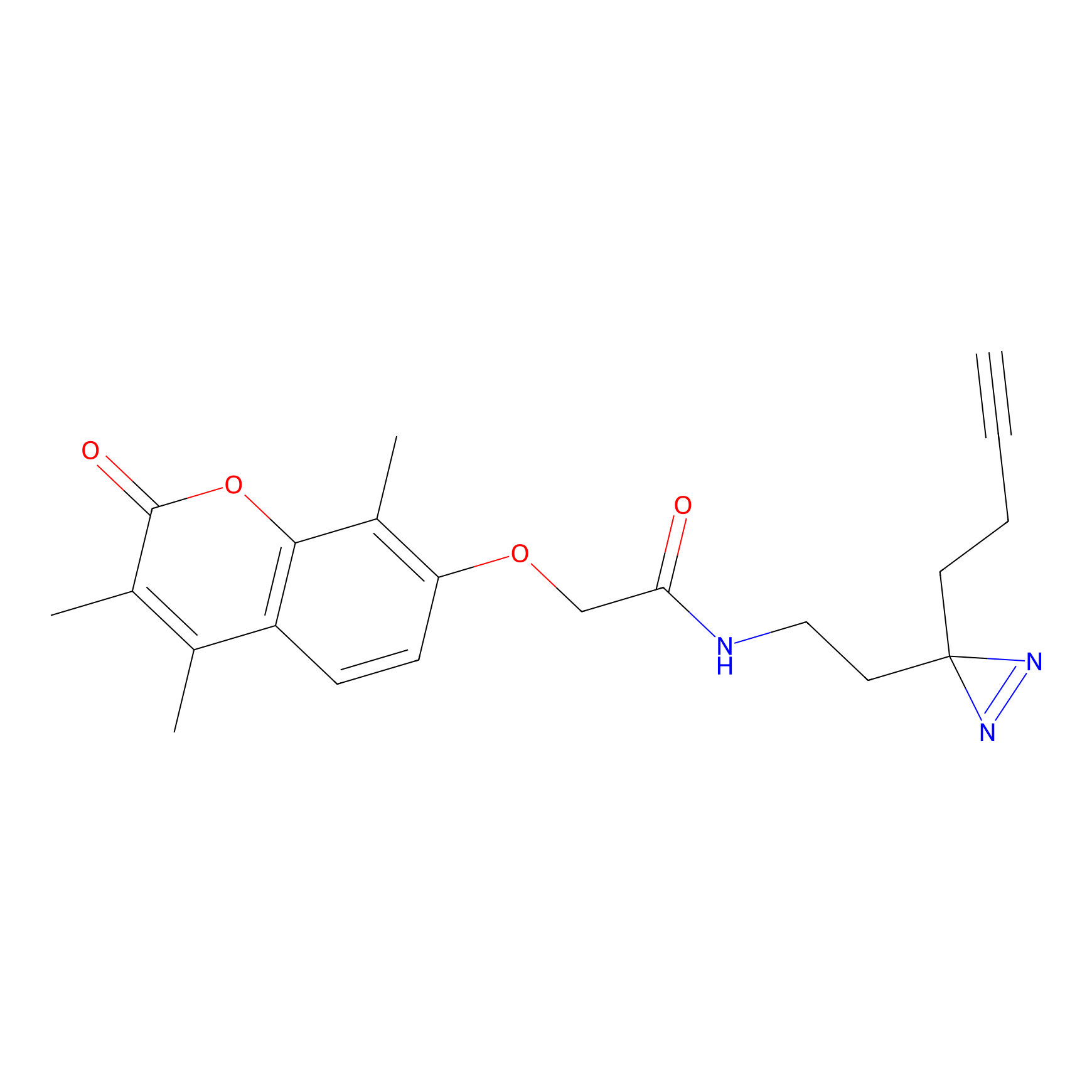 |
12.91 | LDD1713 | [10] | |
|
C094 Probe Info |
 |
27.28 | LDD1785 | [10] | |
|
C112 Probe Info |
 |
18.38 | LDD1799 | [10] | |
|
C158 Probe Info |
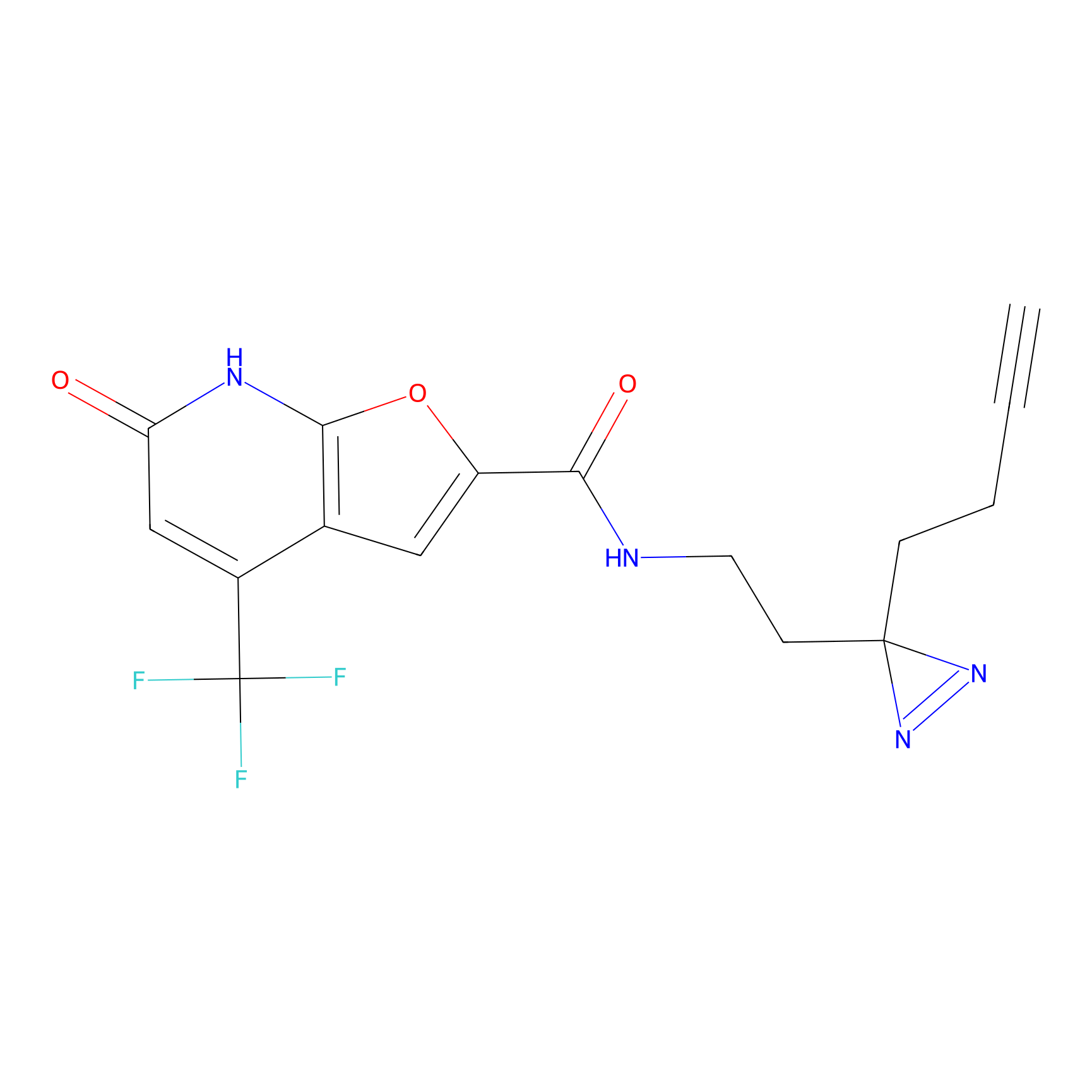 |
9.65 | LDD1838 | [10] | |
|
C210 Probe Info |
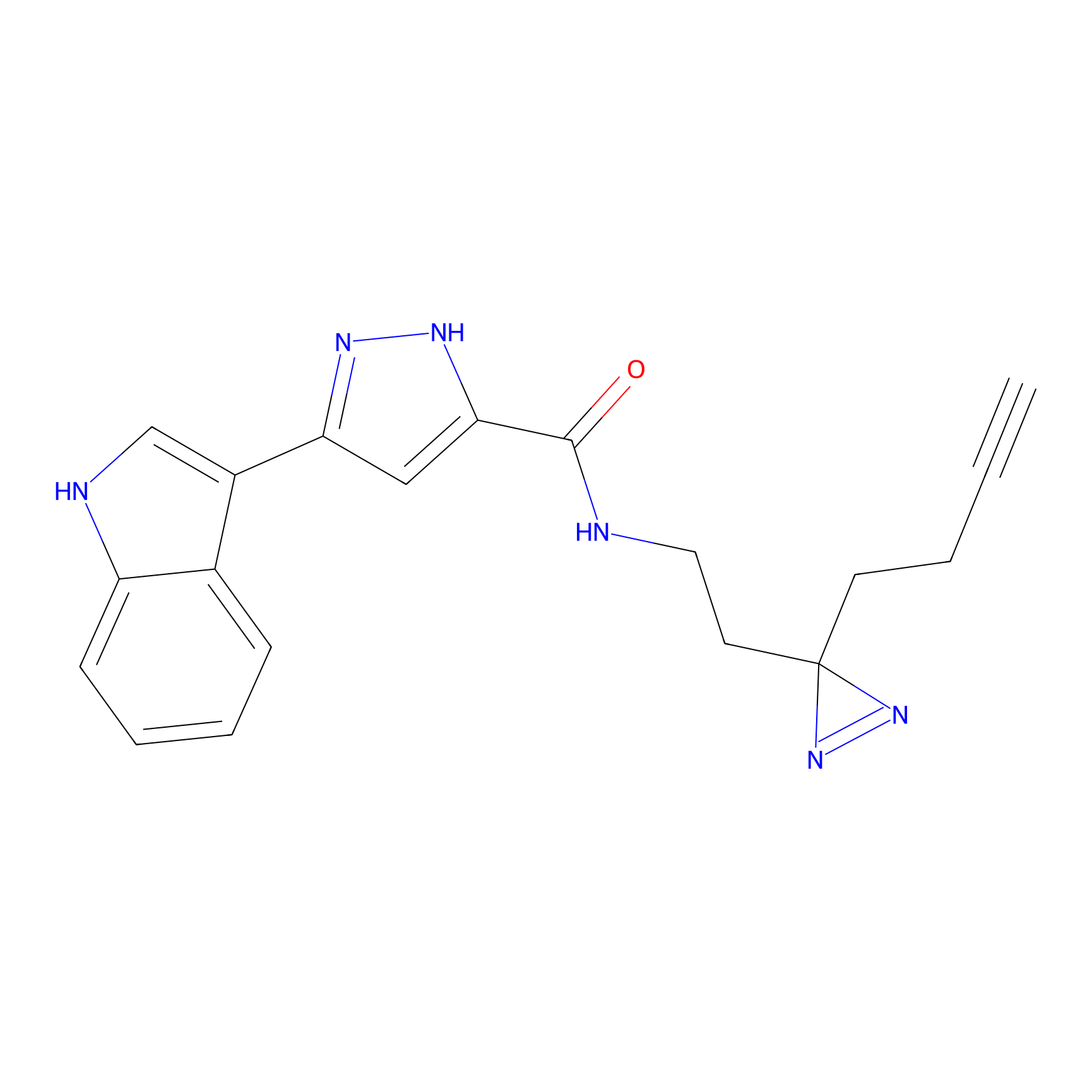 |
51.27 | LDD1884 | [10] | |
|
C355 Probe Info |
 |
25.28 | LDD2016 | [10] | |
|
C361 Probe Info |
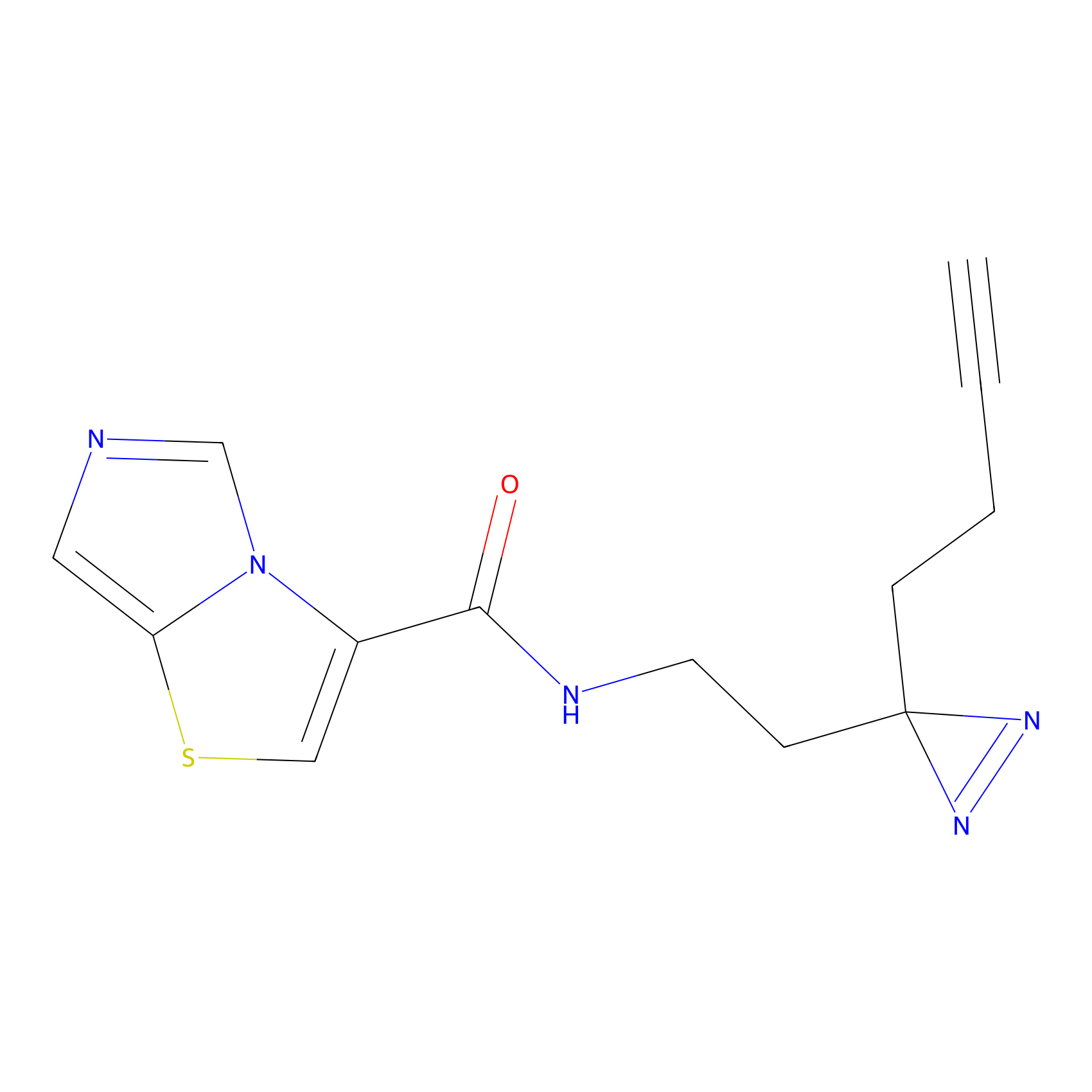 |
18.00 | LDD2022 | [10] | |
Competitor(s) Related to This Target
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Maspardin (SPG21) | AB hydrolase superfamily | Q9NZD8 | |||
| Cytochrome b5 type B (CYB5B) | Cytochrome b5 family | O43169 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Small integral membrane protein 1 (SMIM1) | SMIM1 family | B2RUZ4 | |||
| Sigma intracellular receptor 2 (TMEM97) | TMEM97/sigma-2 receptor family | Q5BJF2 | |||
GPCR
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Lysophosphatidic acid receptor 3 (LPAR3) | G-protein coupled receptor 1 family | Q9UBY5 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Vesicle-trafficking protein SEC22b (SEC22B) | Synaptobrevin family | O75396 | |||
| Syntaxin-8 (STX8) | Syntaxin family | Q9UNK0 | |||
References
