Details of the Target
General Information of Target
| Target ID | LDTP05655 | |||||
|---|---|---|---|---|---|---|
| Target Name | A-kinase anchor protein 13 (AKAP13) | |||||
| Gene Name | AKAP13 | |||||
| Gene ID | 11214 | |||||
| Synonyms |
BRX; HT31; LBC; A-kinase anchor protein 13; AKAP-13; AKAP-Lbc; Breast cancer nuclear receptor-binding auxiliary protein; Guanine nucleotide exchange factor Lbc; Human thyroid-anchoring protein 31; Lymphoid blast crisis oncogene; LBC oncogene; Non-oncogenic Rho GTPase-specific GTP exchange factor; Protein kinase A-anchoring protein 13; PRKA13; p47
|
|||||
| 3D Structure | ||||||
| Sequence |
MKLNPQQAPLYGDCVVTVLLAEEDKAEDDVVFYLVFLGSTLRHCTSTRKVSSDTLETIAP
GHDCCETVKVQLCASKEGLPVFVVAEEDFHFVQDEAYDAAQFLATSAGNQQALNFTRFLD QSGPPSGDVNSLDKKLVLAFRHLKLPTEWNVLGTDQSLHDAGPRETLMHFAVRLGLLRLT WFLLQKPGGRGALSIHNQEGATPVSLALERGYHKLHQLLTEENAGEPDSWSSLSYEIPYG DCSVRHHRELDIYTLTSESDSHHEHPFPGDGCTGPIFKLMNIQQQLMKTNLKQMDSLMPL MMTAQDPSSAPETDGQFLPCAPEPTDPQRLSSSEETESTQCCPGSPVAQTESPCDLSSIV EEENTDRSCRKKNKGVERKGEEVEPAPIVDSGTVSDQDSCLQSLPDCGVKGTEGLSSCGN RNEETGTKSSGMPTDQESLSSGDAVLQRDLVMEPGTAQYSSGGELGGISTTNVSTPDTAG EMEHGLMNPDATVWKNVLQGGESTKERFENSNIGTAGASDVHVTSKPVDKISVPNCAPAA SSLDGNKPAESSLAFSNEETSTEKTAETETSRSREESADAPVDQNSVVIPAAAKDKISDG LEPYTLLAAGIGEAMSPSDLALLGLEEDVMPHQNSETNSSHAQSQKGKSSPICSTTGDDK LCADSACQQNTVTSSGDLVAKLCDNIVSESESTTARQPSSQDPPDASHCEDPQAHTVTSD PVRDTQERADFCPFKVVDNKGQRKDVKLDKPLTNMLEVVSHPHPVVPKMEKELVPDQAVI SDSTFSLANSPGSESVTKDDALSFVPSQKEKGTATPELHTATDYRDGPDGNSNEPDTRPL EDRAVGLSTSSTAAELQHGMGNTSLTGLGGEHEGPAPPAIPEALNIKGNTDSSLQSVGKA TLALDSVLTEEGKLLVVSESSAAQEQDKDKAVTCSSIKENALSSGTLQEEQRTPPPGQDT QQFHEKSISADCAKDKALQLSNSPGASSAFLKAETEHNKEVAPQVSLLTQGGAAQSLVPP GASLATESRQEALGAEHNSSALLPCLLPDGSDGSDALNCSQPSPLDVGVKNTQSQGKTSA CEVSGDVTVDVTGVNALQGMAEPRRENISHNTQDILIPNVLLSQEKNAVLGLPVALQDKA VTDPQGVGTPEMIPLDWEKGKLEGADHSCTMGDAEEAQIDDEAHPVLLQPVAKELPTDME LSAHDDGAPAGVREVMRAPPSGRERSTPSLPCMVSAQDAPLPKGADLIEEAASRIVDAVI EQVKAAGALLTEGEACHMSLSSPELGPLTKGLESAFTEKVSTFPPGESLPMGSTPEEATG SLAGCFAGREEPEKIILPVQGPEPAAEMPDVKAEDEVDFRASSISEEVAVGSIAATLKMK QGPMTQAINRENWCTIEPCPDAASLLASKQSPECENFLDVGLGRECTSKQGVLKRESGSD SDLFHSPSDDMDSIIFPKPEEEHLACDITGSSSSTDDTASLDRHSSHGSDVSLSQILKPN RSRDRQSLDGFYSHGMGAEGRESESEPADPGDVEEEEMDSITEVPANCSVLRSSMRSLSP FRRHSWGPGKNAASDAEMNHRSSMRVLGDVVRRPPIHRRSFSLEGLTGGAGVGNKPSSSL EVSSANAEELRHPFSGEERVDSLVSLSEEDLESDQREHRMFDQQICHRSKQQGFNYCTSA ISSPLTKSISLMTISHPGLDNSRPFHSTFHNTSANLTESITEENYNFLPHSPSKKDSEWK SGTKVSRTFSYIKNKMSSSKKSKEKEKEKDKIKEKEKDSKDKEKDKKTVNGHTFSSIPVV GPISCSQCMKPFTNKDAYTCANCSAFVHKGCRESLASCAKVKMKQPKGSLQAHDTSSLPT VIMRNKPSQPKERPRSAVLLVDETATTPIFANRRSQQSVSLSKSVSIQNITGVGNDENMS NTWKFLSHSTDSLNKISKVNESTESLTDEGVGTDMNEGQLLGDFEIESKQLEAESWSRII DSKFLKQQKKDVVKRQEVIYELMQTEFHHVRTLKIMSGVYSQGMMADLLFEQQMVEKLFP CLDELISIHSQFFQRILERKKESLVDKSEKNFLIKRIGDVLVNQFSGENAERLKKTYGKF CGQHNQSVNYFKDLYAKDKRFQAFVKKKMSSSVVRRLGIPECILLVTQRITKYPVLFQRI LQCTKDNEVEQEDLAQSLSLVKDVIGAVDSKVASYEKKVRLNEIYTKTDSKSIMRMKSGQ MFAKEDLKRKKLVRDGSVFLKNAAGRLKEVQAVLLTDILVFLQEKDQKYIFASLDQKSTV ISLKKLIVREVAHEEKGLFLISMGMTDPEMVEVHASSKEERNSWIQIIQDTINTLNRDED EGIPSENEEEKKMLDTRARELKEQLHQKDQKILLLLEEKEMIFRDMAECSTPLPEDCSPT HSPRVLFRSNTEEALKGGPLMKSAINEVEILQGLVSGNLGGTLGPTVSSPIEQDVVGPVS LPRRAETFGGFDSHQMNASKGGEKEEGDDGQDLRRTESDSGLKKGGNANLVFMLKRNSEQ VVQSVVHLYELLSALQGVVLQQDSYIEDQKLVLSERALTRSLSRPSSLIEQEKQRSLEKQ RQDLANLQKQQAQYLEEKRRREREWEARERELREREALLAQREEEVQQGQQDLEKEREEL QQKKGTYQYDLERLRAAQKQLEREQEQLRREAERLSQRQTERDLCQVSHPHTKLMRIPSF FPSPEEPPSPSAPSIAKSGSLDSELSVSPKRNSISRTHKDKGPFHILSSTSQTNKGPEGQ SQAPASTSASTRLFGLTKPKEKKEKKKKNKTSRSQPGDGPASEVSAEGEEIFC |
|||||
| Target Bioclass |
Other
|
|||||
| Subcellular location |
Cytoplasm, cytosol
|
|||||
| Function |
Scaffold protein that plays an important role in assembling signaling complexes downstream of several types of G protein-coupled receptors. Activates RHOA in response to signaling via G protein-coupled receptors via its function as Rho guanine nucleotide exchange factor. May also activate other Rho family members. Part of a kinase signaling complex that links ADRA1A and ADRA1B adrenergic receptor signaling to the activation of downstream p38 MAP kinases, such as MAPK11 and MAPK14. Part of a signaling complex that links ADRA1B signaling to the activation of RHOA and IKBKB/IKKB, leading to increased NF-kappa-B transcriptional activity. Part of a RHOA-dependent signaling cascade that mediates responses to lysophosphatidic acid (LPA), a signaling molecule that activates G-protein coupled receptors and potentiates transcriptional activation of the glucocorticoid receptor NR3C1. Part of a signaling cascade that stimulates MEF2C-dependent gene expression in response to lysophosphatidic acid (LPA). Part of a signaling pathway that activates MAPK11 and/or MAPK14 and leads to increased transcription activation of the estrogen receptors ESR1 and ESR2. Part of a signaling cascade that links cAMP and EGFR signaling to BRAF signaling and to PKA-mediated phosphorylation of KSR1, leading to the activation of downstream MAP kinases, such as MAPK1 or MAPK3. Functions as a scaffold protein that anchors cAMP-dependent protein kinase (PKA) and PRKD1. This promotes activation of PRKD1, leading to increased phosphorylation of HDAC5 and ultimately cardiomyocyte hypertrophy. Has no guanine nucleotide exchange activity on CDC42, Ras or Rac. Required for normal embryonic heart development, and in particular for normal sarcomere formation in the developing cardiomyocytes. Plays a role in cardiomyocyte growth and cardiac hypertrophy in response to activation of the beta-adrenergic receptor by phenylephrine or isoproterenol. Required for normal adaptive cardiac hypertrophy in response to pressure overload. Plays a role in osteogenesis.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| A498 | SNV: p.A2585S | DBIA Probe Info | |||
| HCT15 | SNV: p.P2419H | DBIA Probe Info | |||
| HEC1 | SNV: p.R1995W | DBIA Probe Info | |||
| HUH1 | SNV: p.L2283S | DBIA Probe Info | |||
| HUH7 | SNV: p.E1261D | DBIA Probe Info | |||
| IGROV1 | Deletion: p.I2372SfsTer10 | DBIA Probe Info | |||
| JHH7 | SNV: p.Q2171R | . | |||
| JURKAT | SNV: p.E1519G; p.R2359Ter | . | |||
| L428 | SNV: p.S944Ter | DBIA Probe Info | |||
| LN18 | SNV: p.Q397P | DBIA Probe Info | |||
| LNCaP clone FGC | SNV: p.Q283R | DBIA Probe Info | |||
| LOXIMVI | SNV: p.D1452G | DBIA Probe Info | |||
| LS180 | SNV: p.A663T | DBIA Probe Info | |||
| MFE319 | SNV: p.D905E | DBIA Probe Info | |||
| NALM6 | SNV: p.E691K; p.R2200C | DBIA Probe Info | |||
| NCIH1703 | SNV: p.Q93Ter | DBIA Probe Info | |||
| NCIH446 | SNV: p.S1453C | DBIA Probe Info | |||
| OVK18 | SNV: p.E502K | DBIA Probe Info | |||
| RKO | Deletion: p.K2504RfsTer50 | DBIA Probe Info | |||
| SKHEP1 | SNV: p.L255S | DBIA Probe Info | |||
| SSP25 | SNV: p.M1538I | DBIA Probe Info | |||
| TE11 | SNV: p.P1150T | DBIA Probe Info | |||
| TOV21G | SNV: p.E2638D | DBIA Probe Info | |||
| U2OS | SNV: p.K2741Ter | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
C-Sul Probe Info |
 |
3.16 | LDD0066 | [2] | |
|
STPyne Probe Info |
 |
K135(10.00) | LDD0277 | [3] | |
|
BTD Probe Info |
 |
C972(0.84) | LDD2099 | [4] | |
|
DA-P3 Probe Info |
 |
28.60 | LDD0179 | [5] | |
|
AHL-Pu-1 Probe Info |
 |
C662(8.27); C536(10.91); C1169(2.83) | LDD0168 | [6] | |
|
DBIA Probe Info |
 |
C732(5.50) | LDD0209 | [7] | |
|
HHS-475 Probe Info |
 |
Y235(1.01) | LDD0264 | [8] | |
|
HHS-465 Probe Info |
 |
Y235(3.58) | LDD2237 | [9] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [10] | |
|
ATP probe Probe Info |
 |
K2191(0.00); K2197(0.00); K2741(0.00) | LDD0199 | [11] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [12] | |
|
IA-alkyne Probe Info |
 |
C44(0.00); C732(0.00); C2142(0.00); C418(0.00) | LDD0162 | [13] | |
|
NAIA_4 Probe Info |
 |
C272(0.00); C536(0.00); C683(0.00); C732(0.00) | LDD2226 | [14] | |
|
WYneN Probe Info |
 |
C2142(0.00); C2163(0.00); C418(0.00); C732(0.00) | LDD0021 | [15] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [15] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [15] | |
|
TFBX Probe Info |
 |
C732(0.00); C1232(0.00); C2101(0.00); C44(0.00) | LDD0148 | [16] | |
|
VSF Probe Info |
 |
C683(0.00); C1232(0.00) | LDD0007 | [15] | |
|
YN-1 Probe Info |
 |
N.A. | LDD0447 | [17] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [18] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [19] | |
|
AOyne Probe Info |
 |
14.00 | LDD0443 | [20] | |
|
NAIA_5 Probe Info |
 |
C2101(0.00); C418(0.00); C14(0.00) | LDD2223 | [14] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
Photocelecoxib Probe Info |
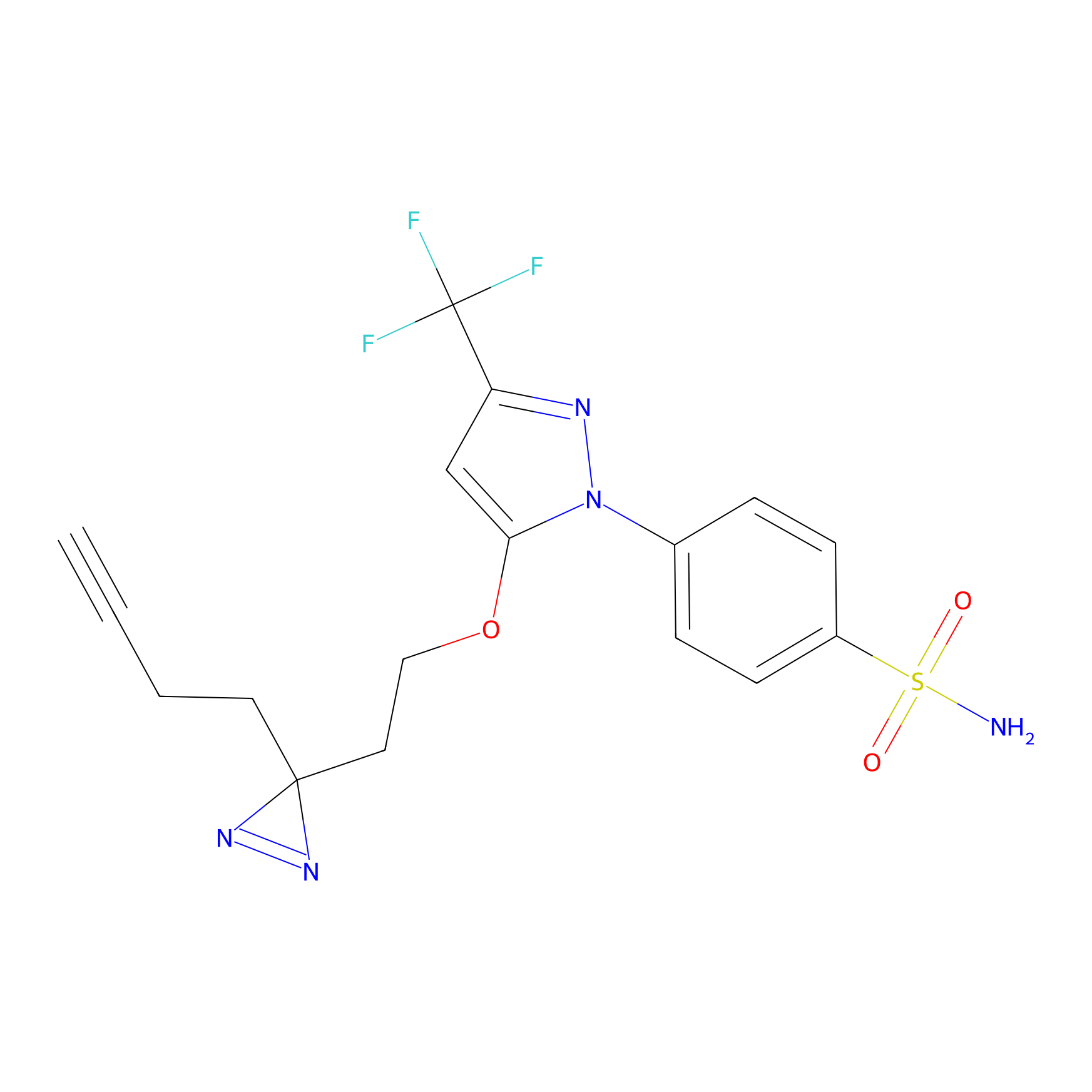 |
N.A. | LDD0019 | [21] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | HEK-293T | C662(8.27); C536(10.91); C1169(2.83) | LDD0168 | [6] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C709(3.32); C662(10.71); C400(2.20) | LDD0169 | [6] |
| LDCM0214 | AC1 | HEK-293T | C972(0.87); C934(1.03); C683(0.79); C2142(1.09) | LDD1507 | [22] |
| LDCM0215 | AC10 | HEK-293T | C972(0.93); C934(0.94); C683(1.06); C2142(1.00) | LDD1508 | [22] |
| LDCM0226 | AC11 | HEK-293T | C972(0.90); C934(0.98); C683(0.90); C2142(1.03) | LDD1509 | [22] |
| LDCM0237 | AC12 | HEK-293T | C972(0.93); C934(1.02); C683(0.91); C2142(0.91) | LDD1510 | [22] |
| LDCM0259 | AC14 | HEK-293T | C972(0.98); C934(1.03); C683(1.04); C2142(1.04) | LDD1512 | [22] |
| LDCM0270 | AC15 | HEK-293T | C972(0.96); C934(1.04); C683(1.02); C1276(1.04) | LDD1513 | [22] |
| LDCM0276 | AC17 | HEK-293T | C972(1.10); C934(1.10); C683(0.92); C2142(0.99) | LDD1515 | [22] |
| LDCM0277 | AC18 | HEK-293T | C972(0.99); C934(1.06); C683(1.09); C2142(0.87) | LDD1516 | [22] |
| LDCM0278 | AC19 | HEK-293T | C972(1.27); C934(1.42); C683(1.83); C2142(1.06) | LDD1517 | [22] |
| LDCM0279 | AC2 | HEK-293T | C972(0.89); C934(0.93); C683(0.98); C2142(0.96) | LDD1518 | [22] |
| LDCM0280 | AC20 | HEK-293T | C972(1.00); C934(1.07); C683(0.95); C2142(0.92) | LDD1519 | [22] |
| LDCM0281 | AC21 | HEK-293T | C972(1.04); C934(1.01); C683(1.11); C2142(0.96) | LDD1520 | [22] |
| LDCM0282 | AC22 | HEK-293T | C972(1.03); C934(1.11); C683(1.00); C2142(1.06) | LDD1521 | [22] |
| LDCM0283 | AC23 | HEK-293T | C972(1.03); C934(1.06); C683(1.03); C1276(1.07) | LDD1522 | [22] |
| LDCM0284 | AC24 | HEK-293T | C972(1.16); C934(1.04); C683(1.09); C2142(1.03) | LDD1523 | [22] |
| LDCM0285 | AC25 | HEK-293T | C972(1.00); C934(1.06); C683(0.97); C2142(0.88) | LDD1524 | [22] |
| LDCM0286 | AC26 | HEK-293T | C972(0.89); C934(0.99); C683(0.77); C2142(0.96) | LDD1525 | [22] |
| LDCM0287 | AC27 | HEK-293T | C972(0.94); C934(0.99); C683(1.19); C2142(1.13) | LDD1526 | [22] |
| LDCM0288 | AC28 | HEK-293T | C972(0.95); C934(1.05); C683(0.86); C2142(0.98) | LDD1527 | [22] |
| LDCM0289 | AC29 | HEK-293T | C972(0.99); C934(1.05); C683(1.00); C2142(1.06) | LDD1528 | [22] |
| LDCM0290 | AC3 | HEK-293T | C972(0.90); C934(0.96); C683(0.82); C2142(1.10) | LDD1529 | [22] |
| LDCM0291 | AC30 | HEK-293T | C972(0.95); C934(1.00); C683(1.09); C2142(0.92) | LDD1530 | [22] |
| LDCM0292 | AC31 | HEK-293T | C972(0.95); C934(1.07); C683(1.10); C1276(1.10) | LDD1531 | [22] |
| LDCM0293 | AC32 | HEK-293T | C972(1.04); C934(1.04); C683(1.56); C2142(0.98) | LDD1532 | [22] |
| LDCM0294 | AC33 | HEK-293T | C972(1.08); C934(1.27); C683(0.96); C2142(0.87) | LDD1533 | [22] |
| LDCM0295 | AC34 | HEK-293T | C972(0.99); C934(1.03); C683(0.90); C2142(0.99) | LDD1534 | [22] |
| LDCM0296 | AC35 | HEK-293T | C972(0.97); C934(1.10); C683(1.18); C2142(1.10) | LDD1535 | [22] |
| LDCM0297 | AC36 | HEK-293T | C972(1.01); C934(1.15); C683(1.07); C2142(0.99) | LDD1536 | [22] |
| LDCM0298 | AC37 | HEK-293T | C972(1.00); C934(0.90); C683(1.06); C2142(1.07) | LDD1537 | [22] |
| LDCM0299 | AC38 | HEK-293T | C972(0.95); C934(1.08); C683(0.80); C2142(0.98) | LDD1538 | [22] |
| LDCM0300 | AC39 | HEK-293T | C972(0.96); C934(1.12); C683(0.93); C1276(1.20) | LDD1539 | [22] |
| LDCM0301 | AC4 | HEK-293T | C972(0.85); C934(0.93); C683(0.98); C2142(0.96) | LDD1540 | [22] |
| LDCM0302 | AC40 | HEK-293T | C972(1.03); C934(1.01); C683(0.98); C2142(1.00) | LDD1541 | [22] |
| LDCM0303 | AC41 | HEK-293T | C972(0.98); C934(1.16); C683(1.04); C2142(0.89) | LDD1542 | [22] |
| LDCM0304 | AC42 | HEK-293T | C972(0.98); C934(1.01); C683(0.86); C2142(1.13) | LDD1543 | [22] |
| LDCM0305 | AC43 | HEK-293T | C972(0.94); C934(1.13); C683(0.86); C2142(1.05) | LDD1544 | [22] |
| LDCM0306 | AC44 | HEK-293T | C972(1.02); C934(1.11); C683(0.99); C2142(1.04) | LDD1545 | [22] |
| LDCM0307 | AC45 | HEK-293T | C972(1.10); C934(0.97); C683(1.02); C2142(1.01) | LDD1546 | [22] |
| LDCM0308 | AC46 | HEK-293T | C972(1.05); C934(1.06); C683(0.88); C2142(1.01) | LDD1547 | [22] |
| LDCM0309 | AC47 | HEK-293T | C972(0.99); C934(1.02); C683(0.92); C1276(1.13) | LDD1548 | [22] |
| LDCM0310 | AC48 | HEK-293T | C972(1.09); C934(1.11); C683(1.19); C2142(1.02) | LDD1549 | [22] |
| LDCM0311 | AC49 | HEK-293T | C972(0.95); C934(1.04); C683(1.01); C2142(1.01) | LDD1550 | [22] |
| LDCM0312 | AC5 | HEK-293T | C972(1.02); C934(0.92); C683(1.01); C2142(1.03) | LDD1551 | [22] |
| LDCM0313 | AC50 | HEK-293T | C972(0.92); C934(0.96); C683(0.86); C2142(0.90) | LDD1552 | [22] |
| LDCM0314 | AC51 | HEK-293T | C972(0.91); C934(1.02); C683(0.74); C2142(1.18) | LDD1553 | [22] |
| LDCM0315 | AC52 | HEK-293T | C972(0.95); C934(1.09); C683(0.95); C2142(1.00) | LDD1554 | [22] |
| LDCM0316 | AC53 | HEK-293T | C972(1.07); C934(1.03); C683(0.95); C2142(1.04) | LDD1555 | [22] |
| LDCM0317 | AC54 | HEK-293T | C972(1.06); C934(1.14); C683(0.81); C2142(0.96) | LDD1556 | [22] |
| LDCM0318 | AC55 | HEK-293T | C972(1.07); C934(1.04); C683(1.19); C1276(1.10) | LDD1557 | [22] |
| LDCM0319 | AC56 | HEK-293T | C972(1.12); C934(1.08); C683(1.15); C2142(1.04) | LDD1558 | [22] |
| LDCM0320 | AC57 | HEK-293T | C972(0.90); C934(1.06); C683(1.15); C2142(0.97) | LDD1559 | [22] |
| LDCM0321 | AC58 | HEK-293T | C972(0.92); C934(0.96); C683(1.06); C2142(0.99) | LDD1560 | [22] |
| LDCM0322 | AC59 | HEK-293T | C972(0.89); C934(1.02); C683(1.19); C2142(1.02) | LDD1561 | [22] |
| LDCM0323 | AC6 | HEK-293T | C972(0.97); C934(0.99); C683(0.75); C2142(0.98) | LDD1562 | [22] |
| LDCM0324 | AC60 | HEK-293T | C972(0.93); C934(1.01); C683(1.04); C2142(1.02) | LDD1563 | [22] |
| LDCM0325 | AC61 | HEK-293T | C972(1.08); C934(0.99); C683(1.03); C2142(1.02) | LDD1564 | [22] |
| LDCM0326 | AC62 | HEK-293T | C972(1.00); C934(1.12); C683(0.87); C2142(0.96) | LDD1565 | [22] |
| LDCM0327 | AC63 | HEK-293T | C972(1.04); C934(1.05); C683(1.13); C1276(1.05) | LDD1566 | [22] |
| LDCM0328 | AC64 | HEK-293T | C972(1.04); C934(0.98); C683(0.81); C2142(0.91) | LDD1567 | [22] |
| LDCM0334 | AC7 | HEK-293T | C972(0.92); C934(0.95); C683(0.84); C1276(1.10) | LDD1568 | [22] |
| LDCM0345 | AC8 | HEK-293T | C972(1.01); C934(0.97); C683(1.13); C2142(0.96) | LDD1569 | [22] |
| LDCM0248 | AKOS034007472 | HEK-293T | C972(1.06); C934(1.02); C683(1.01); C2142(0.96) | LDD1511 | [22] |
| LDCM0356 | AKOS034007680 | HEK-293T | C972(0.95); C934(1.04); C683(1.11); C2142(0.99) | LDD1570 | [22] |
| LDCM0275 | AKOS034007705 | HEK-293T | C972(1.01); C934(0.97); C683(1.01); C2142(1.02) | LDD1514 | [22] |
| LDCM0156 | Aniline | NCI-H1299 | 11.76 | LDD0403 | [1] |
| LDCM0108 | Chloroacetamide | HeLa | C1232(0.00); C732(0.00) | LDD0222 | [19] |
| LDCM0632 | CL-Sc | Hep-G2 | C418(20.00); C1232(2.20); C2101(0.82); C2101(0.75) | LDD2227 | [14] |
| LDCM0367 | CL1 | HEK-293T | C972(0.82); C934(0.87); C683(1.12); C1276(1.06) | LDD1571 | [22] |
| LDCM0368 | CL10 | HEK-293T | C972(0.84); C934(0.84); C683(0.85); C2142(1.22) | LDD1572 | [22] |
| LDCM0369 | CL100 | HEK-293T | C972(1.00); C934(0.98); C683(1.15); C2142(1.12) | LDD1573 | [22] |
| LDCM0370 | CL101 | HEK-293T | C972(1.03); C934(1.05); C683(0.99); C1276(1.20) | LDD1574 | [22] |
| LDCM0371 | CL102 | HEK-293T | C972(0.92); C934(0.93); C683(1.03); C1276(1.05) | LDD1575 | [22] |
| LDCM0372 | CL103 | HEK-293T | C972(1.01); C934(1.10); C683(1.05); C2142(0.97) | LDD1576 | [22] |
| LDCM0373 | CL104 | HEK-293T | C972(0.98); C934(0.95); C683(0.89); C2142(1.06) | LDD1577 | [22] |
| LDCM0374 | CL105 | HEK-293T | C972(1.01); C934(1.09); C683(1.11); C1276(1.03) | LDD1578 | [22] |
| LDCM0375 | CL106 | HEK-293T | C972(0.99); C934(1.03); C683(0.97); C1276(1.04) | LDD1579 | [22] |
| LDCM0376 | CL107 | HEK-293T | C972(1.01); C934(1.11); C683(1.07); C2142(0.97) | LDD1580 | [22] |
| LDCM0377 | CL108 | HEK-293T | C972(1.09); C934(1.09); C683(1.21); C2142(0.93) | LDD1581 | [22] |
| LDCM0378 | CL109 | HEK-293T | C972(0.93); C934(1.03); C683(0.92); C1276(0.98) | LDD1582 | [22] |
| LDCM0379 | CL11 | HEK-293T | C972(0.92); C934(0.89); C683(1.17); C1276(1.18) | LDD1583 | [22] |
| LDCM0380 | CL110 | HEK-293T | C972(0.91); C934(0.94); C683(0.95); C1276(0.88) | LDD1584 | [22] |
| LDCM0381 | CL111 | HEK-293T | C972(0.83); C934(1.05); C683(0.96); C2142(0.94) | LDD1585 | [22] |
| LDCM0382 | CL112 | HEK-293T | C972(0.98); C934(1.04); C683(1.28); C2142(1.20) | LDD1586 | [22] |
| LDCM0383 | CL113 | HEK-293T | C972(0.98); C934(1.15); C683(0.98); C1276(0.98) | LDD1587 | [22] |
| LDCM0384 | CL114 | HEK-293T | C972(0.89); C934(0.94); C683(1.02); C1276(0.97) | LDD1588 | [22] |
| LDCM0385 | CL115 | HEK-293T | C972(0.93); C934(1.11); C683(0.98); C2142(0.92) | LDD1589 | [22] |
| LDCM0386 | CL116 | HEK-293T | C972(1.00); C934(1.13); C683(0.92); C2142(1.03) | LDD1590 | [22] |
| LDCM0387 | CL117 | HEK-293T | C972(1.03); C934(1.13); C683(1.05); C1276(1.02) | LDD1591 | [22] |
| LDCM0388 | CL118 | HEK-293T | C972(0.97); C934(1.04); C683(1.03); C1276(1.00) | LDD1592 | [22] |
| LDCM0389 | CL119 | HEK-293T | C972(0.93); C934(1.03); C683(1.10); C2142(0.94) | LDD1593 | [22] |
| LDCM0390 | CL12 | HEK-293T | C972(1.02); C934(0.94); C683(1.34); C2142(1.08) | LDD1594 | [22] |
| LDCM0391 | CL120 | HEK-293T | C972(0.95); C934(1.04); C683(1.12); C2142(1.05) | LDD1595 | [22] |
| LDCM0392 | CL121 | HEK-293T | C972(0.99); C934(1.06); C683(1.07); C1276(1.06) | LDD1596 | [22] |
| LDCM0393 | CL122 | HEK-293T | C972(0.94); C934(1.00); C683(1.06); C1276(1.06) | LDD1597 | [22] |
| LDCM0394 | CL123 | HEK-293T | C972(0.89); C934(1.01); C683(1.15); C2142(1.05) | LDD1598 | [22] |
| LDCM0395 | CL124 | HEK-293T | C972(0.92); C934(1.03); C683(1.09); C2142(1.18) | LDD1599 | [22] |
| LDCM0396 | CL125 | HEK-293T | C972(0.99); C934(1.01); C683(1.05); C1276(0.88) | LDD1600 | [22] |
| LDCM0397 | CL126 | HEK-293T | C972(1.00); C934(1.07); C683(1.12); C1276(1.11) | LDD1601 | [22] |
| LDCM0398 | CL127 | HEK-293T | C972(0.91); C934(1.06); C683(1.11); C2142(0.97) | LDD1602 | [22] |
| LDCM0399 | CL128 | HEK-293T | C972(1.05); C934(1.04); C683(1.20); C2142(1.19) | LDD1603 | [22] |
| LDCM0400 | CL13 | HEK-293T | C972(0.97); C934(1.02); C683(1.12); C1276(1.13) | LDD1604 | [22] |
| LDCM0401 | CL14 | HEK-293T | C972(0.93); C934(0.95); C683(1.01); C1276(0.96) | LDD1605 | [22] |
| LDCM0402 | CL15 | HEK-293T | C972(0.88); C934(0.97); C683(1.21); C2142(1.08) | LDD1606 | [22] |
| LDCM0403 | CL16 | HEK-293T | C972(0.97); C934(0.99); C683(1.30); C2142(1.03) | LDD1607 | [22] |
| LDCM0404 | CL17 | HEK-293T | C972(0.95); C934(0.96); C683(0.99); C2142(1.11) | LDD1608 | [22] |
| LDCM0405 | CL18 | HEK-293T | C972(0.88); C934(0.98); C683(1.19); C2142(1.01) | LDD1609 | [22] |
| LDCM0406 | CL19 | HEK-293T | C972(0.98); C934(1.00); C683(1.06); C2142(1.34) | LDD1610 | [22] |
| LDCM0407 | CL2 | HEK-293T | C972(0.91); C934(0.93); C683(1.06); C1276(1.04) | LDD1611 | [22] |
| LDCM0408 | CL20 | HEK-293T | C972(0.96); C934(1.04); C683(1.11); C2142(1.03) | LDD1612 | [22] |
| LDCM0409 | CL21 | HEK-293T | C972(1.13); C934(0.99); C683(1.04); C2142(1.24) | LDD1613 | [22] |
| LDCM0410 | CL22 | HEK-293T | C972(0.98); C934(0.98); C683(0.88); C2142(1.15) | LDD1614 | [22] |
| LDCM0411 | CL23 | HEK-293T | C972(0.95); C934(1.00); C683(1.04); C1276(1.11) | LDD1615 | [22] |
| LDCM0412 | CL24 | HEK-293T | C972(1.05); C934(1.04); C683(1.02); C2142(1.06) | LDD1616 | [22] |
| LDCM0413 | CL25 | HEK-293T | C972(0.97); C934(1.06); C683(0.99); C1276(0.86) | LDD1617 | [22] |
| LDCM0414 | CL26 | HEK-293T | C972(1.02); C934(1.07); C683(0.93); C1276(1.01) | LDD1618 | [22] |
| LDCM0415 | CL27 | HEK-293T | C972(0.95); C934(1.07); C683(1.12); C2142(0.91) | LDD1619 | [22] |
| LDCM0416 | CL28 | HEK-293T | C972(1.12); C934(0.97); C683(1.33); C2142(1.06) | LDD1620 | [22] |
| LDCM0417 | CL29 | HEK-293T | C972(0.95); C934(1.01); C683(0.89); C2142(0.84) | LDD1621 | [22] |
| LDCM0418 | CL3 | HEK-293T | C972(0.82); C934(0.90); C683(1.07); C2142(1.06) | LDD1622 | [22] |
| LDCM0419 | CL30 | HEK-293T | C972(0.97); C934(0.94); C683(0.97); C2142(0.82) | LDD1623 | [22] |
| LDCM0420 | CL31 | HEK-293T | C972(0.95); C934(1.01); C683(1.27); C2142(1.15) | LDD1624 | [22] |
| LDCM0421 | CL32 | HEK-293T | C972(0.97); C934(1.05); C683(0.98); C2142(1.05) | LDD1625 | [22] |
| LDCM0422 | CL33 | HEK-293T | C972(1.02); C934(0.89); C683(0.93); C2142(0.99) | LDD1626 | [22] |
| LDCM0423 | CL34 | HEK-293T | C972(1.01); C934(1.06); C683(0.95); C2142(1.14) | LDD1627 | [22] |
| LDCM0424 | CL35 | HEK-293T | C972(1.01); C934(1.09); C683(1.05); C1276(1.15) | LDD1628 | [22] |
| LDCM0425 | CL36 | HEK-293T | C972(1.14); C934(1.07); C683(0.85); C2142(0.97) | LDD1629 | [22] |
| LDCM0426 | CL37 | HEK-293T | C972(1.02); C934(1.08); C683(1.03); C1276(0.90) | LDD1630 | [22] |
| LDCM0428 | CL39 | HEK-293T | C972(1.07); C934(1.02); C683(1.04); C2142(0.99) | LDD1632 | [22] |
| LDCM0429 | CL4 | HEK-293T | C972(0.96); C934(0.89); C683(1.12); C2142(1.12) | LDD1633 | [22] |
| LDCM0430 | CL40 | HEK-293T | C972(1.07); C934(1.04); C683(1.90); C2142(1.07) | LDD1634 | [22] |
| LDCM0431 | CL41 | HEK-293T | C972(1.04); C934(1.01); C683(1.03); C2142(0.97) | LDD1635 | [22] |
| LDCM0432 | CL42 | HEK-293T | C972(0.99); C934(0.94); C683(0.99); C2142(1.00) | LDD1636 | [22] |
| LDCM0433 | CL43 | HEK-293T | C972(0.99); C934(0.95); C683(1.11); C2142(1.01) | LDD1637 | [22] |
| LDCM0434 | CL44 | HEK-293T | C972(1.00); C934(1.08); C683(1.10); C2142(1.06) | LDD1638 | [22] |
| LDCM0435 | CL45 | HEK-293T | C972(1.17); C934(0.98); C683(0.92); C2142(1.12) | LDD1639 | [22] |
| LDCM0436 | CL46 | HEK-293T | C972(1.19); C934(1.09); C683(1.00); C2142(1.03) | LDD1640 | [22] |
| LDCM0437 | CL47 | HEK-293T | C972(1.01); C934(1.08); C683(1.00); C1276(1.06) | LDD1641 | [22] |
| LDCM0438 | CL48 | HEK-293T | C972(1.18); C934(1.16); C683(1.08); C2142(0.97) | LDD1642 | [22] |
| LDCM0439 | CL49 | HEK-293T | C972(0.88); C934(0.97); C683(1.00); C1276(0.86) | LDD1643 | [22] |
| LDCM0440 | CL5 | HEK-293T | C972(0.95); C934(0.95); C683(0.84); C2142(0.95) | LDD1644 | [22] |
| LDCM0441 | CL50 | HEK-293T | C972(0.95); C934(0.99); C683(1.02); C1276(1.14) | LDD1645 | [22] |
| LDCM0443 | CL52 | HEK-293T | C972(0.94); C934(0.98); C683(0.98); C2142(1.09) | LDD1646 | [22] |
| LDCM0444 | CL53 | HEK-293T | C972(0.86); C934(1.00); C683(1.00); C2142(1.11) | LDD1647 | [22] |
| LDCM0445 | CL54 | HEK-293T | C972(0.81); C934(0.83); C683(0.95); C2142(1.01) | LDD1648 | [22] |
| LDCM0446 | CL55 | HEK-293T | C972(0.95); C934(1.06); C683(1.03); C2142(1.20) | LDD1649 | [22] |
| LDCM0447 | CL56 | HEK-293T | C972(0.94); C934(1.06); C683(1.21); C2142(1.05) | LDD1650 | [22] |
| LDCM0448 | CL57 | HEK-293T | C972(1.22); C934(0.96); C683(1.29); C2142(1.08) | LDD1651 | [22] |
| LDCM0449 | CL58 | HEK-293T | C972(0.93); C934(1.04); C683(0.99); C2142(1.17) | LDD1652 | [22] |
| LDCM0450 | CL59 | HEK-293T | C972(1.02); C934(1.00); C683(1.08); C1276(1.07) | LDD1653 | [22] |
| LDCM0451 | CL6 | HEK-293T | C972(0.81); C934(0.88); C683(0.93); C2142(1.04) | LDD1654 | [22] |
| LDCM0452 | CL60 | HEK-293T | C972(1.20); C934(1.07); C683(1.60); C2142(0.98) | LDD1655 | [22] |
| LDCM0453 | CL61 | HEK-293T | C972(0.99); C934(1.09); C683(1.08); C1276(0.87) | LDD1656 | [22] |
| LDCM0454 | CL62 | HEK-293T | C972(1.00); C934(1.04); C683(0.99); C1276(0.99) | LDD1657 | [22] |
| LDCM0455 | CL63 | HEK-293T | C972(0.99); C934(1.13); C683(1.23); C2142(0.88) | LDD1658 | [22] |
| LDCM0456 | CL64 | HEK-293T | C972(0.92); C934(1.03); C683(1.10); C2142(1.29) | LDD1659 | [22] |
| LDCM0457 | CL65 | HEK-293T | C972(0.99); C934(1.15); C683(1.18); C2142(1.09) | LDD1660 | [22] |
| LDCM0458 | CL66 | HEK-293T | C972(0.98); C934(1.03); C683(1.28); C2142(1.01) | LDD1661 | [22] |
| LDCM0459 | CL67 | HEK-293T | C972(0.96); C934(0.98); C683(1.36); C2142(1.27) | LDD1662 | [22] |
| LDCM0460 | CL68 | HEK-293T | C972(0.99); C934(1.06); C683(0.98); C2142(1.11) | LDD1663 | [22] |
| LDCM0461 | CL69 | HEK-293T | C972(1.18); C934(0.98); C683(1.09); C2142(1.17) | LDD1664 | [22] |
| LDCM0462 | CL7 | HEK-293T | C972(0.90); C934(1.00); C683(1.22); C2142(1.13) | LDD1665 | [22] |
| LDCM0463 | CL70 | HEK-293T | C972(1.05); C934(1.10); C683(0.90); C2142(1.09) | LDD1666 | [22] |
| LDCM0464 | CL71 | HEK-293T | C972(1.01); C934(1.03); C683(1.03); C1276(1.05) | LDD1667 | [22] |
| LDCM0465 | CL72 | HEK-293T | C972(1.42); C934(1.19); C683(1.35); C2142(1.01) | LDD1668 | [22] |
| LDCM0466 | CL73 | HEK-293T | C972(0.89); C934(0.91); C683(0.89); C1276(0.91) | LDD1669 | [22] |
| LDCM0467 | CL74 | HEK-293T | C972(0.88); C934(0.94); C683(1.08); C1276(1.04) | LDD1670 | [22] |
| LDCM0469 | CL76 | HEK-293T | C972(1.03); C934(0.98); C683(1.18); C2142(1.07) | LDD1672 | [22] |
| LDCM0470 | CL77 | HEK-293T | C972(0.90); C934(0.82); C683(0.91); C2142(1.05) | LDD1673 | [22] |
| LDCM0471 | CL78 | HEK-293T | C972(0.91); C934(0.93); C683(0.99); C2142(1.14) | LDD1674 | [22] |
| LDCM0472 | CL79 | HEK-293T | C972(0.93); C934(0.92); C683(1.32); C2142(1.18) | LDD1675 | [22] |
| LDCM0473 | CL8 | HEK-293T | C972(0.73); C934(0.76); C683(0.86); C2142(1.39) | LDD1676 | [22] |
| LDCM0474 | CL80 | HEK-293T | C972(0.91); C934(1.00); C683(1.04); C2142(0.98) | LDD1677 | [22] |
| LDCM0475 | CL81 | HEK-293T | C972(1.16); C934(1.04); C683(1.03); C2142(1.08) | LDD1678 | [22] |
| LDCM0476 | CL82 | HEK-293T | C972(1.04); C934(1.00); C683(0.99); C2142(1.00) | LDD1679 | [22] |
| LDCM0477 | CL83 | HEK-293T | C972(0.87); C934(0.91); C683(0.96); C1276(1.16) | LDD1680 | [22] |
| LDCM0478 | CL84 | HEK-293T | C972(1.01); C934(0.97); C683(0.90); C2142(1.01) | LDD1681 | [22] |
| LDCM0479 | CL85 | HEK-293T | C972(0.91); C934(1.01); C683(0.96); C1276(0.90) | LDD1682 | [22] |
| LDCM0480 | CL86 | HEK-293T | C972(0.97); C934(1.05); C683(0.96); C1276(0.89) | LDD1683 | [22] |
| LDCM0481 | CL87 | HEK-293T | C972(0.92); C934(1.12); C683(1.01); C2142(0.88) | LDD1684 | [22] |
| LDCM0482 | CL88 | HEK-293T | C972(0.98); C934(1.09); C683(1.03); C2142(0.99) | LDD1685 | [22] |
| LDCM0483 | CL89 | HEK-293T | C972(0.96); C934(1.08); C683(0.96); C2142(0.90) | LDD1686 | [22] |
| LDCM0484 | CL9 | HEK-293T | C972(1.02); C934(0.85); C683(0.91); C2142(1.12) | LDD1687 | [22] |
| LDCM0485 | CL90 | HEK-293T | C972(0.83); C934(0.80); C683(0.92); C2142(1.06) | LDD1688 | [22] |
| LDCM0486 | CL91 | HEK-293T | C972(0.92); C934(1.00); C683(1.21); C2142(1.04) | LDD1689 | [22] |
| LDCM0487 | CL92 | HEK-293T | C972(0.99); C934(1.07); C683(1.03); C2142(1.08) | LDD1690 | [22] |
| LDCM0488 | CL93 | HEK-293T | C972(1.11); C934(0.95); C683(1.36); C2142(1.18) | LDD1691 | [22] |
| LDCM0489 | CL94 | HEK-293T | C972(1.16); C934(1.11); C683(0.77); C2142(1.12) | LDD1692 | [22] |
| LDCM0490 | CL95 | HEK-293T | C972(0.96); C934(0.99); C683(1.04); C1276(1.19) | LDD1693 | [22] |
| LDCM0491 | CL96 | HEK-293T | C972(1.12); C934(1.00); C683(0.95); C2142(0.94) | LDD1694 | [22] |
| LDCM0492 | CL97 | HEK-293T | C972(0.92); C934(1.03); C683(1.07); C1276(0.96) | LDD1695 | [22] |
| LDCM0493 | CL98 | HEK-293T | C972(0.95); C934(0.97); C683(1.04); C1276(1.11) | LDD1696 | [22] |
| LDCM0494 | CL99 | HEK-293T | C972(0.90); C934(1.06); C683(1.12); C2142(0.95) | LDD1697 | [22] |
| LDCM0634 | CY-0357 | Hep-G2 | C2101(0.86) | LDD2228 | [14] |
| LDCM0028 | Dobutamine | HEK-293T | 9.11 | LDD0180 | [5] |
| LDCM0027 | Dopamine | HEK-293T | 28.60 | LDD0179 | [5] |
| LDCM0495 | E2913 | HEK-293T | C972(1.09); C934(1.20); C683(1.09); C2142(0.93) | LDD1698 | [22] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C732(1.70) | LDD1702 | [4] |
| LDCM0625 | F8 | Ramos | C732(0.74); C1232(1.28) | LDD2187 | [23] |
| LDCM0572 | Fragment10 | Ramos | C732(2.08) | LDD2189 | [23] |
| LDCM0573 | Fragment11 | Ramos | C732(20.00) | LDD2190 | [23] |
| LDCM0574 | Fragment12 | Ramos | C732(1.45); C1232(1.23) | LDD2191 | [23] |
| LDCM0575 | Fragment13 | Ramos | C732(1.15); C1232(1.07) | LDD2192 | [23] |
| LDCM0576 | Fragment14 | Ramos | C732(1.28); C1232(0.90) | LDD2193 | [23] |
| LDCM0579 | Fragment20 | Ramos | C732(1.22); C1232(1.80) | LDD2194 | [23] |
| LDCM0580 | Fragment21 | Ramos | C732(1.08); C1232(0.82) | LDD2195 | [23] |
| LDCM0582 | Fragment23 | Ramos | C732(1.40); C1232(1.17) | LDD2196 | [23] |
| LDCM0578 | Fragment27 | Ramos | C732(1.03); C1232(0.95) | LDD2197 | [23] |
| LDCM0586 | Fragment28 | Ramos | C732(0.83); C1232(0.70) | LDD2198 | [23] |
| LDCM0588 | Fragment30 | Ramos | C732(1.53); C1232(1.51) | LDD2199 | [23] |
| LDCM0589 | Fragment31 | Ramos | C732(1.15); C1232(1.25) | LDD2200 | [23] |
| LDCM0590 | Fragment32 | Ramos | C732(6.17) | LDD2201 | [23] |
| LDCM0468 | Fragment33 | HEK-293T | C972(0.82); C934(0.94); C683(1.07); C2142(0.92) | LDD1671 | [22] |
| LDCM0596 | Fragment38 | Ramos | C732(1.19); C1232(1.07) | LDD2203 | [23] |
| LDCM0566 | Fragment4 | Ramos | C732(1.61); C1232(1.18) | LDD2184 | [23] |
| LDCM0427 | Fragment51 | HEK-293T | C972(1.04); C934(1.07); C683(1.07); C1276(0.92) | LDD1631 | [22] |
| LDCM0610 | Fragment52 | Ramos | C732(1.37); C1232(1.20) | LDD2204 | [23] |
| LDCM0614 | Fragment56 | Ramos | C732(1.19); C1232(1.29) | LDD2205 | [23] |
| LDCM0569 | Fragment7 | Ramos | C732(1.59); C1232(1.00) | LDD2186 | [23] |
| LDCM0571 | Fragment9 | Ramos | C732(1.49) | LDD2188 | [23] |
| LDCM0116 | HHS-0101 | DM93 | Y235(1.01) | LDD0264 | [8] |
| LDCM0117 | HHS-0201 | DM93 | Y235(1.06) | LDD0265 | [8] |
| LDCM0118 | HHS-0301 | DM93 | Y235(1.34) | LDD0266 | [8] |
| LDCM0119 | HHS-0401 | DM93 | Y235(1.30) | LDD0267 | [8] |
| LDCM0120 | HHS-0701 | DM93 | Y235(1.06) | LDD0268 | [8] |
| LDCM0022 | KB02 | HEK-293T | C709(1.15); C418(0.94); C2685(0.91); C972(0.99) | LDD1492 | [22] |
| LDCM0023 | KB03 | Jurkat | C732(5.50) | LDD0209 | [7] |
| LDCM0024 | KB05 | COLO792 | C2689(1.95); C418(1.42) | LDD3310 | [24] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0231 | [19] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C972(0.84) | LDD2099 | [4] |
| LDCM0029 | Quercetin | HEK-293T | 14.31 | LDD0181 | [5] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Protein-glutamine gamma-glutamyltransferase 2 (TGM2) | Transglutaminase family | P21980 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| 14-3-3 protein zeta/delta (YWHAZ) | 14-3-3 family | P63104 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Estrogen receptor (ESR1) | Nuclear hormone receptor family | P03372 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Microtubule-associated proteins 1A/1B light chain 3A (MAP1LC3A) | ATG8 family | Q9H492 | |||
References
