Details of the Target
General Information of Target
| Target ID | LDTP05089 | |||||
|---|---|---|---|---|---|---|
| Target Name | Immunoglobulin-binding protein 1 (IGBP1) | |||||
| Gene Name | IGBP1 | |||||
| Gene ID | 3476 | |||||
| Synonyms |
IBP1; Immunoglobulin-binding protein 1; B-cell signal transduction molecule alpha 4; Protein alpha-4; CD79a-binding protein 1; Protein phosphatase 2/4/6 regulatory subunit; Renal carcinoma antigen NY-REN-16
|
|||||
| 3D Structure | ||||||
| Sequence |
MAAEDELQLPRLPELFETGRQLLDEVEVATEPAGSRIVQEKVFKGLDLLEKAAEMLSQLD
LFSRNEDLEEIASTDLKYLLVPAFQGALTMKQVNPSKRLDHLQRAREHFINYLTQCHCYH VAEFELPKTMNNSAENHTANSSMAYPSLVAMASQRQAKIQRYKQKKELEHRLSAMKSAVE SGQADDERVREYYLLHLQRWIDISLEEIESIDQEIKILRERDSSREASTSNSSRQERPPV KPFILTRNMAQAKVFGAGYPSLPTMTVSDWYEQHRKYGALPDQGIAKAAPEEFRKAAQQQ EEQEEKEEEDDEQTLHRAREWDDWKDTHPRGYGNRQNMG |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
IGBP1/TAP42 family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Associated to surface IgM-receptor; may be involved in signal transduction. Involved in regulation of the catalytic activity of the phosphatases PP2A, PP4 and PP6 by protecting their partially folded catalytic subunits from degradative polyubiquitination until they associate with regulatory subunits.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
STPyne Probe Info |
 |
K166(10.00); K241(2.13); K276(5.36); K44(1.10) | LDD0277 | [2] | |
|
Probe 1 Probe Info |
 |
Y277(17.15) | LDD3495 | [3] | |
|
HHS-475 Probe Info |
 |
Y259(1.00); Y271(1.23) | LDD0264 | [4] | |
|
HHS-465 Probe Info |
 |
Y277(8.55) | LDD2237 | [5] | |
|
DBIA Probe Info |
 |
C118(1.05) | LDD1510 | [6] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [7] | |
|
Ox-W18 Probe Info |
 |
W321(0.00); W324(0.00) | LDD2175 | [8] | |
|
AOyne Probe Info |
 |
14.50 | LDD0443 | [9] | |
|
HHS-482 Probe Info |
 |
Y259(1.32); Y271(1.12); Y277(1.24) | LDD2239 | [5] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C017 Probe Info |
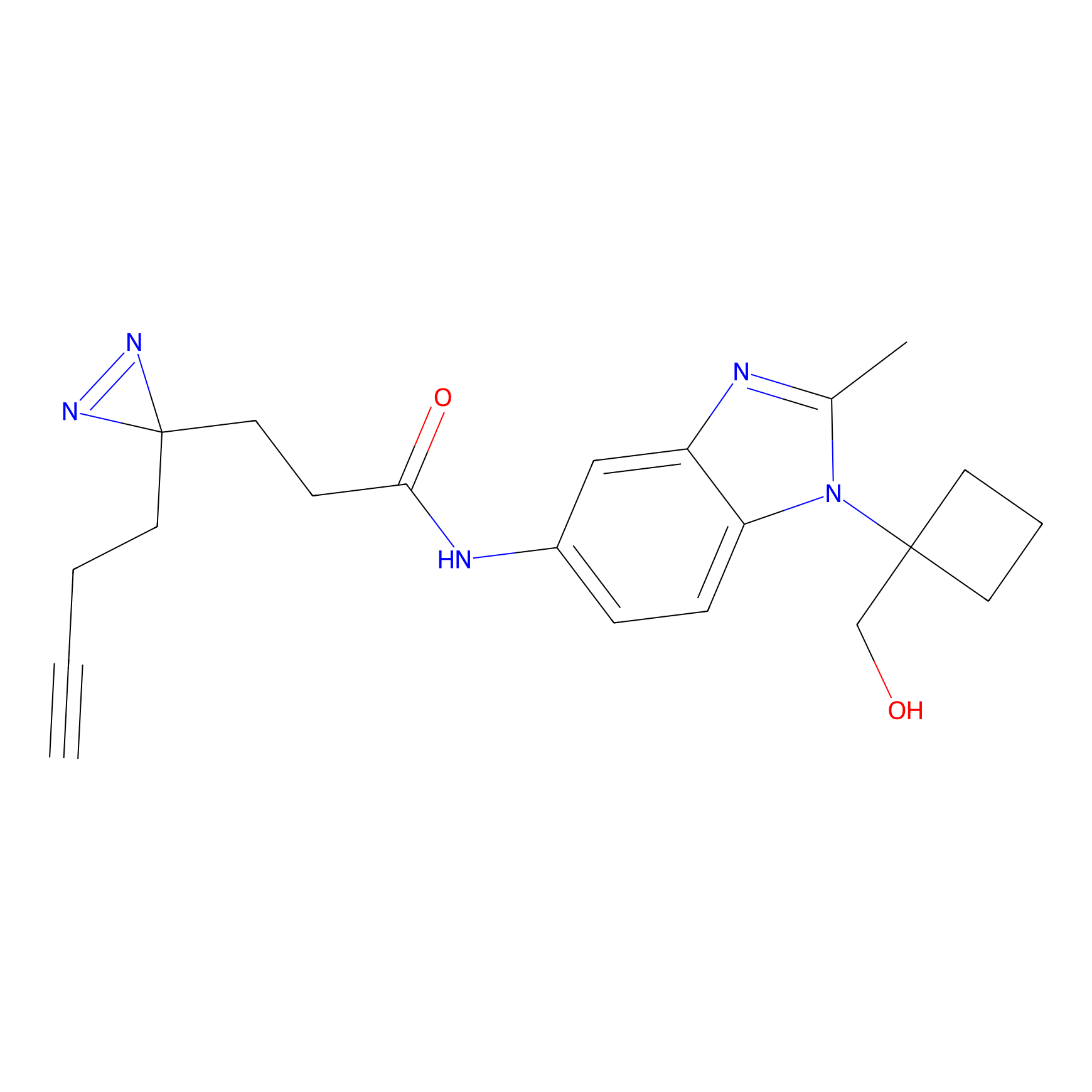 |
7.94 | LDD1725 | [10] | |
|
C158 Probe Info |
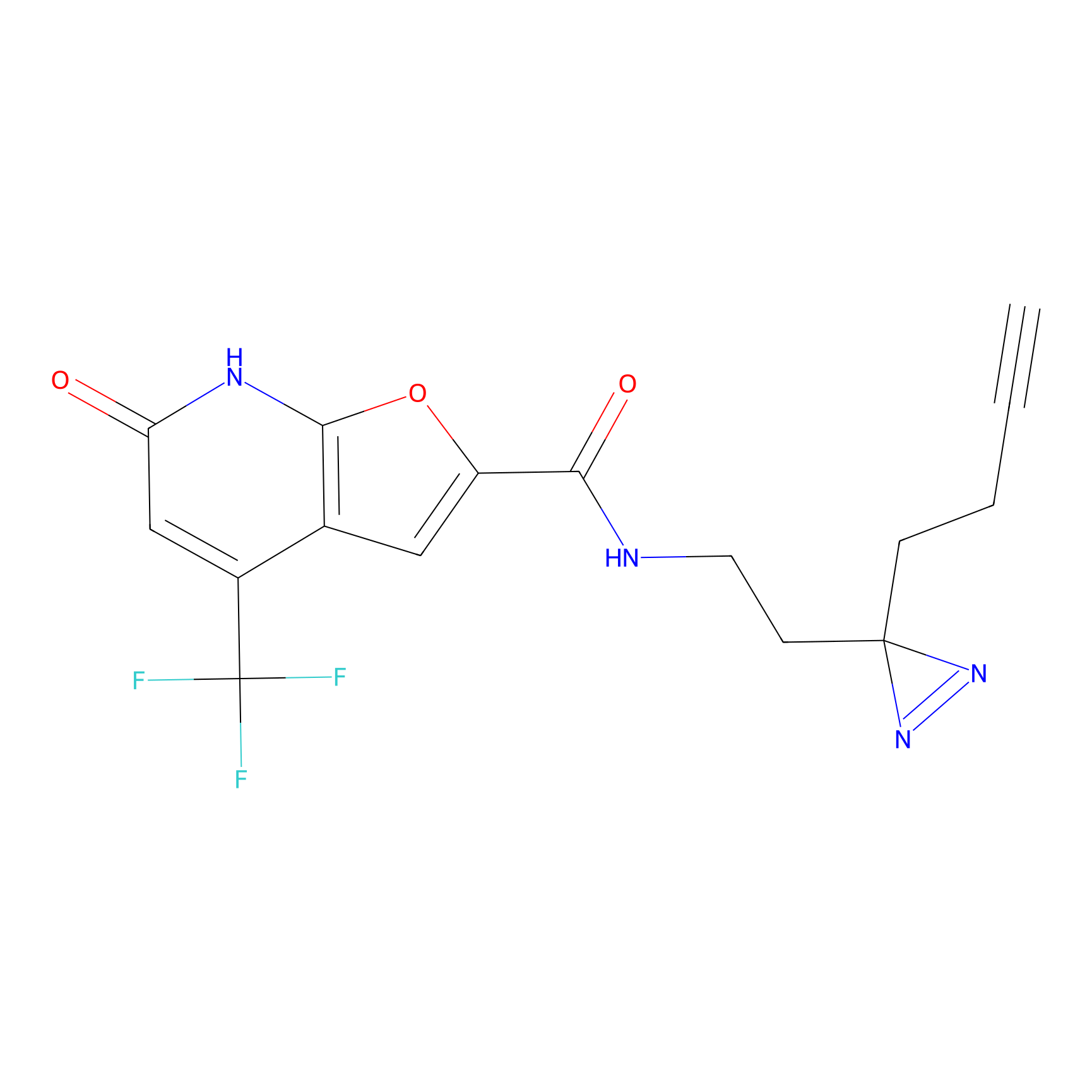 |
13.83 | LDD1838 | [10] | |
|
C252 Probe Info |
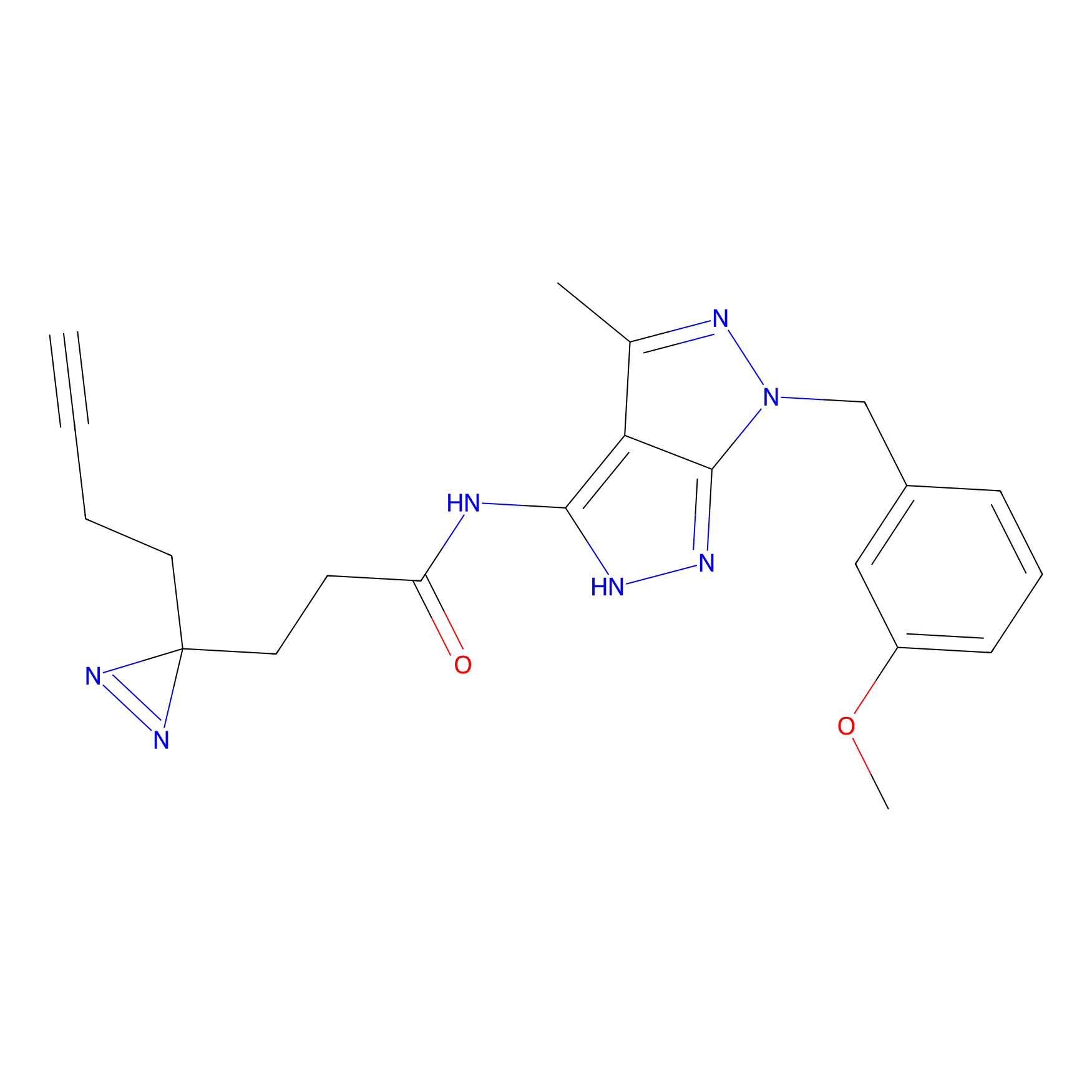 |
10.63 | LDD1925 | [10] | |
|
C313 Probe Info |
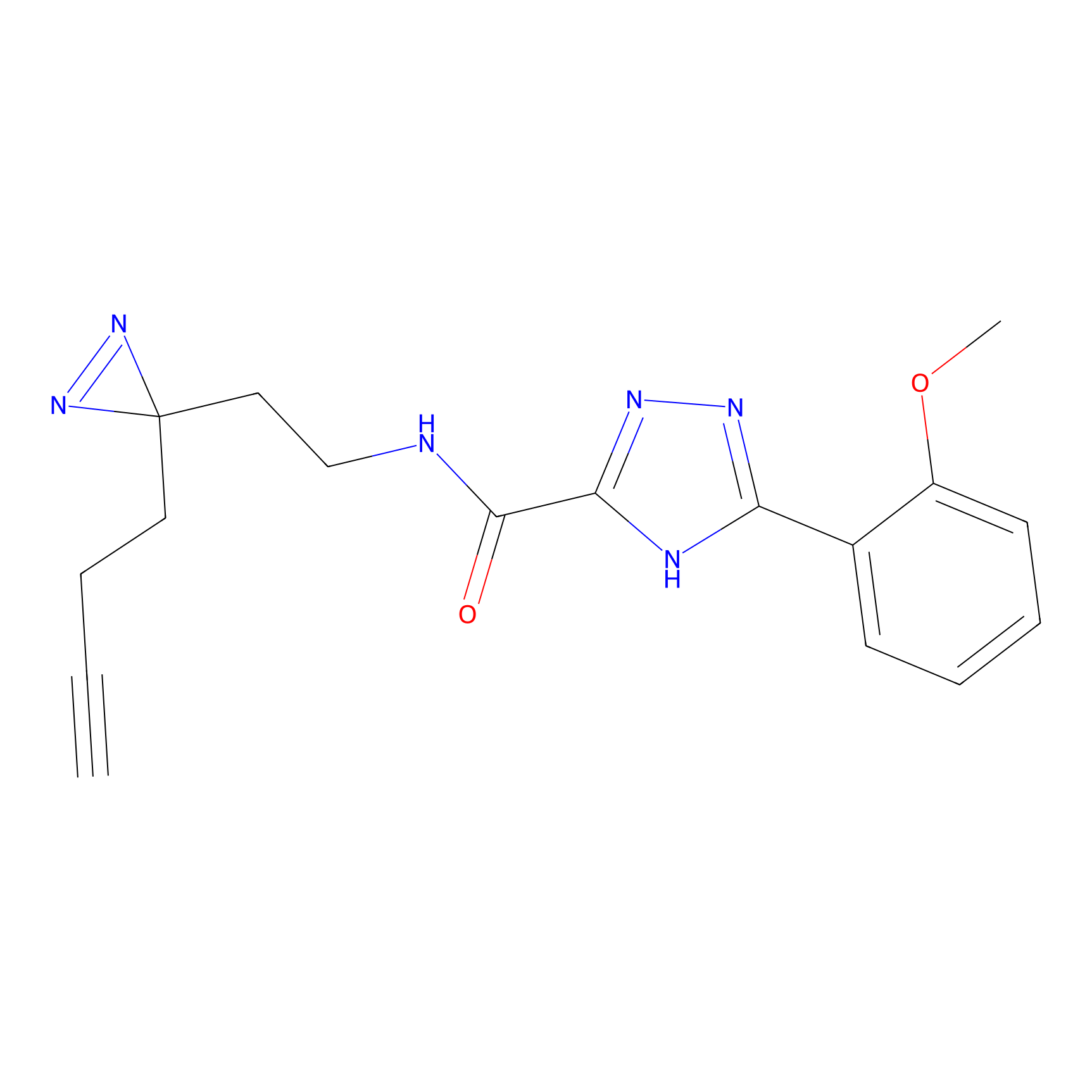 |
13.55 | LDD1980 | [10] | |
|
C317 Probe Info |
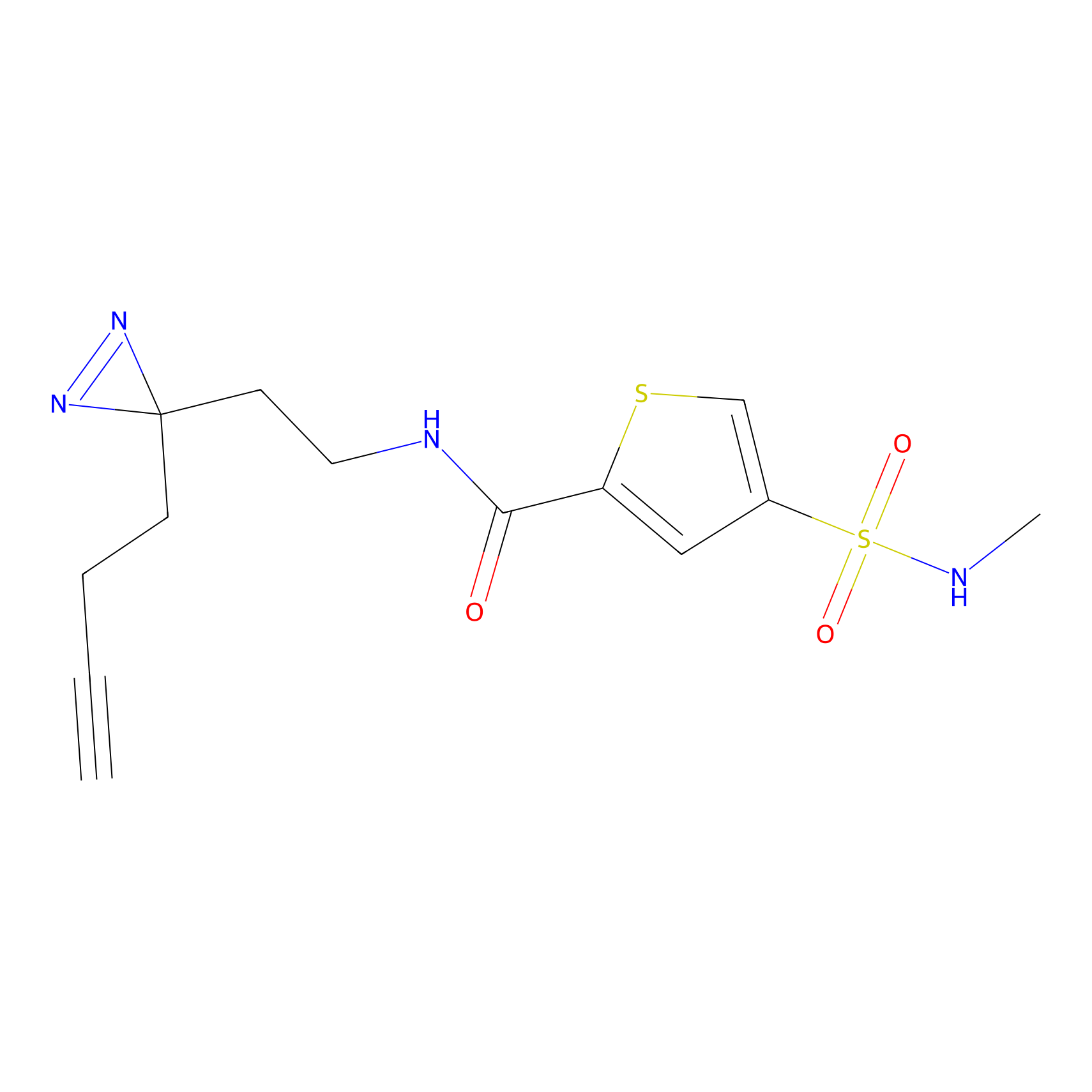 |
6.06 | LDD1983 | [10] | |
|
C363 Probe Info |
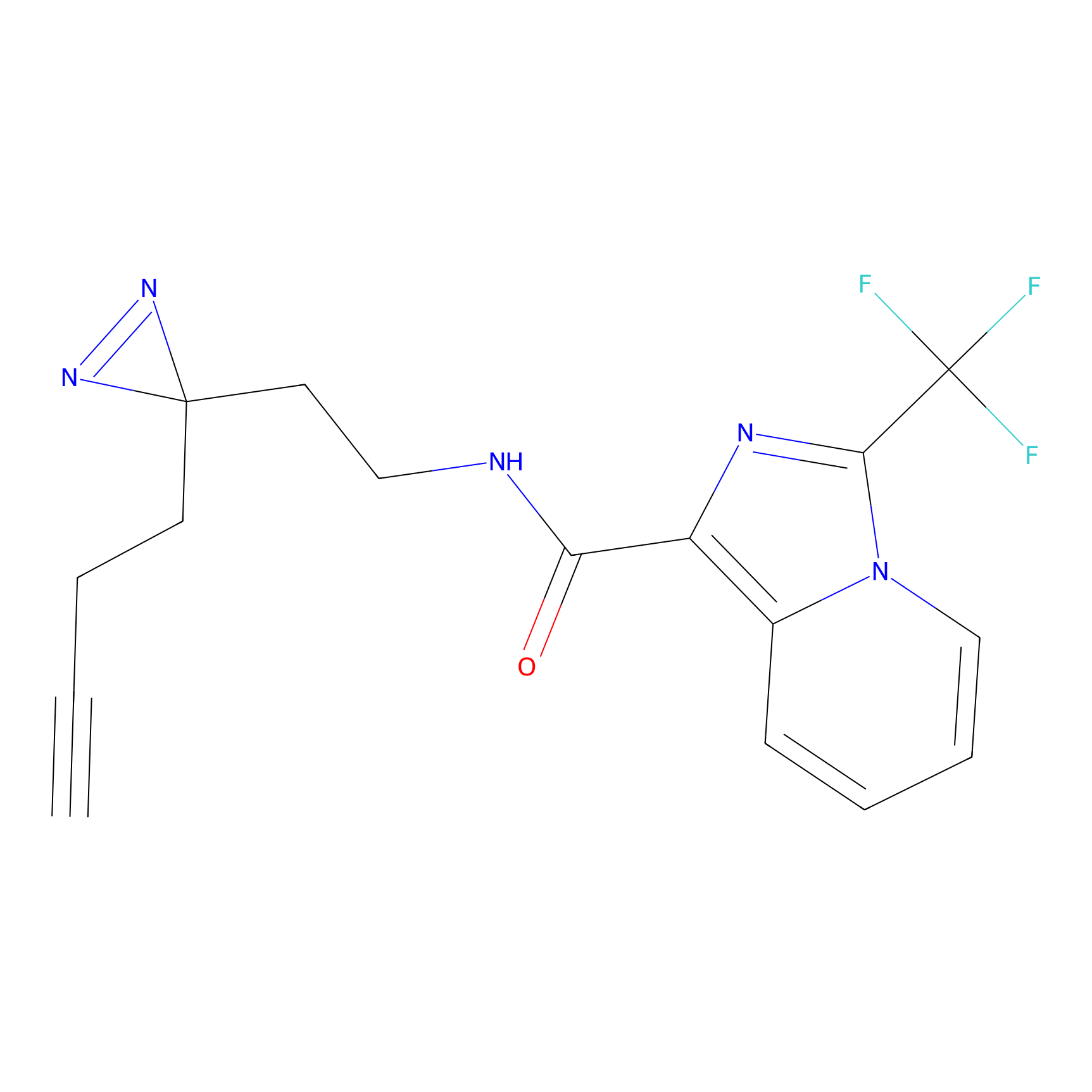 |
16.56 | LDD2024 | [10] | |
|
C407 Probe Info |
 |
14.52 | LDD2064 | [10] | |
|
C419 Probe Info |
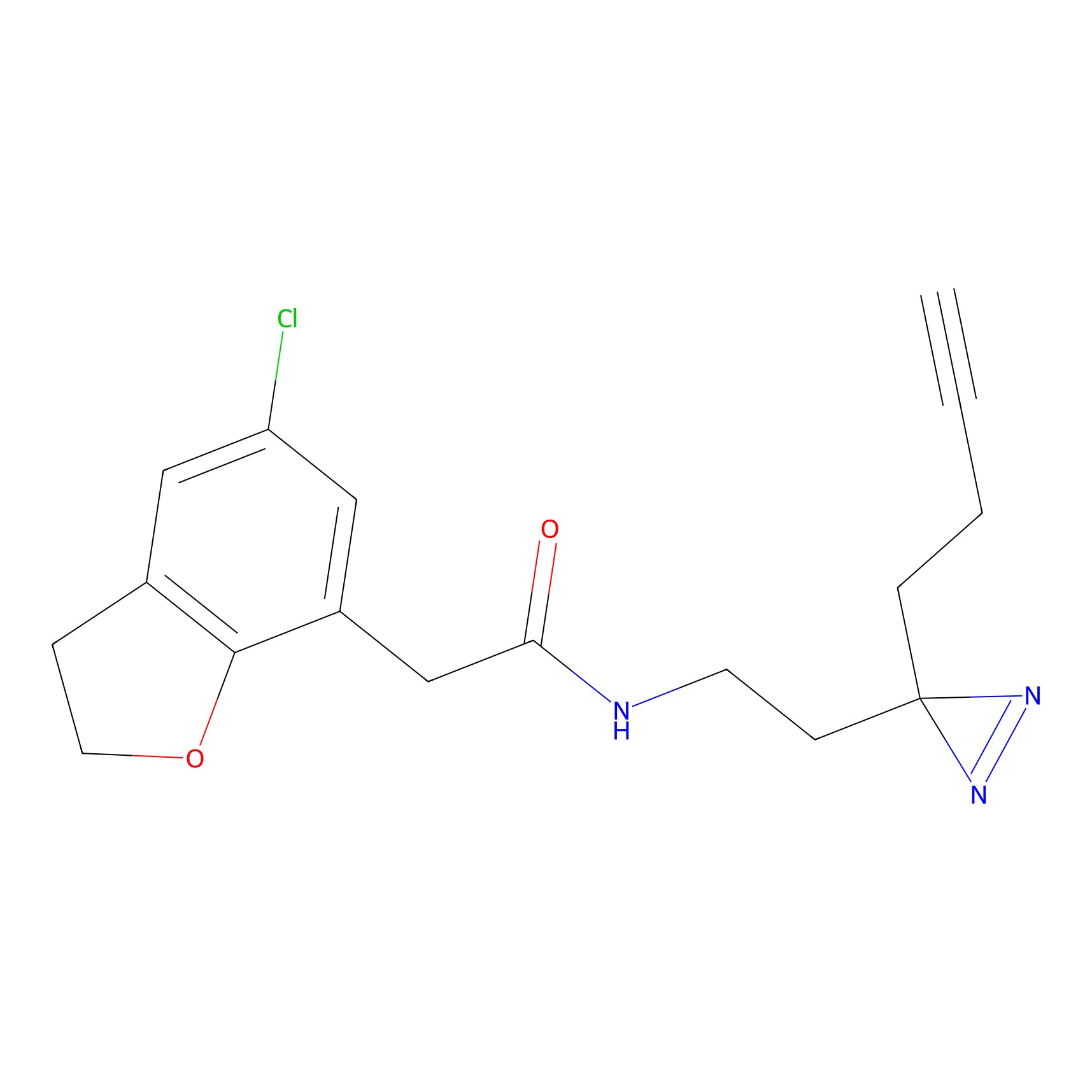 |
17.15 | LDD2074 | [10] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0237 | AC12 | HEK-293T | C118(1.05) | LDD1510 | [6] |
| LDCM0280 | AC20 | HEK-293T | C118(0.96) | LDD1519 | [6] |
| LDCM0288 | AC28 | HEK-293T | C118(0.91) | LDD1527 | [6] |
| LDCM0297 | AC36 | HEK-293T | C118(1.09) | LDD1536 | [6] |
| LDCM0301 | AC4 | HEK-293T | C118(0.98) | LDD1540 | [6] |
| LDCM0306 | AC44 | HEK-293T | C118(1.04) | LDD1545 | [6] |
| LDCM0315 | AC52 | HEK-293T | C118(0.93) | LDD1554 | [6] |
| LDCM0324 | AC60 | HEK-293T | C118(0.97) | LDD1563 | [6] |
| LDCM0156 | Aniline | NCI-H1299 | 15.00 | LDD0403 | [1] |
| LDCM0372 | CL103 | HEK-293T | C118(1.23) | LDD1576 | [6] |
| LDCM0376 | CL107 | HEK-293T | C118(1.07) | LDD1580 | [6] |
| LDCM0381 | CL111 | HEK-293T | C118(1.42) | LDD1585 | [6] |
| LDCM0385 | CL115 | HEK-293T | C118(1.13) | LDD1589 | [6] |
| LDCM0389 | CL119 | HEK-293T | C118(1.22) | LDD1593 | [6] |
| LDCM0394 | CL123 | HEK-293T | C118(1.30) | LDD1598 | [6] |
| LDCM0398 | CL127 | HEK-293T | C118(1.21) | LDD1602 | [6] |
| LDCM0402 | CL15 | HEK-293T | C118(1.18) | LDD1606 | [6] |
| LDCM0408 | CL20 | HEK-293T | C118(1.01) | LDD1612 | [6] |
| LDCM0415 | CL27 | HEK-293T | C118(1.02) | LDD1619 | [6] |
| LDCM0418 | CL3 | HEK-293T | C118(1.37) | LDD1622 | [6] |
| LDCM0421 | CL32 | HEK-293T | C118(0.89) | LDD1625 | [6] |
| LDCM0428 | CL39 | HEK-293T | C118(1.24) | LDD1632 | [6] |
| LDCM0434 | CL44 | HEK-293T | C118(0.86) | LDD1638 | [6] |
| LDCM0447 | CL56 | HEK-293T | C118(0.99) | LDD1650 | [6] |
| LDCM0455 | CL63 | HEK-293T | C118(1.32) | LDD1658 | [6] |
| LDCM0460 | CL68 | HEK-293T | C118(0.92) | LDD1663 | [6] |
| LDCM0473 | CL8 | HEK-293T | C118(0.93) | LDD1676 | [6] |
| LDCM0474 | CL80 | HEK-293T | C118(0.91) | LDD1677 | [6] |
| LDCM0481 | CL87 | HEK-293T | C118(1.21) | LDD1684 | [6] |
| LDCM0487 | CL92 | HEK-293T | C118(0.90) | LDD1690 | [6] |
| LDCM0494 | CL99 | HEK-293T | C118(1.11) | LDD1697 | [6] |
| LDCM0495 | E2913 | HEK-293T | C118(1.67) | LDD1698 | [6] |
| LDCM0468 | Fragment33 | HEK-293T | C118(1.13) | LDD1671 | [6] |
| LDCM0116 | HHS-0101 | DM93 | Y259(1.00); Y271(1.23) | LDD0264 | [4] |
| LDCM0117 | HHS-0201 | DM93 | Y259(0.73); Y271(0.74) | LDD0265 | [4] |
| LDCM0118 | HHS-0301 | DM93 | Y259(0.11); Y271(0.48) | LDD0266 | [4] |
| LDCM0119 | HHS-0401 | DM93 | Y259(0.25); Y271(0.88) | LDD0267 | [4] |
| LDCM0120 | HHS-0701 | DM93 | Y259(1.22); Y271(1.33) | LDD0268 | [4] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| 14-3-3 protein gamma (YWHAG) | 14-3-3 family | P61981 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Heat shock protein beta-1 (HSPB1) | Small heat shock protein (HSP20) family | P04792 | |||
| TIP41-like protein (TIPRL) | TIP41 family | O75663 | |||
References
