Details of the Target
General Information of Target
| Target ID | LDTP01616 | |||||
|---|---|---|---|---|---|---|
| Target Name | CTD nuclear envelope phosphatase 1 (CTDNEP1) | |||||
| Gene Name | CTDNEP1 | |||||
| Gene ID | 23399 | |||||
| Synonyms |
DULLARD; CTD nuclear envelope phosphatase 1; EC 3.1.3.16; Serine/threonine-protein phosphatase dullard |
|||||
| 3D Structure | ||||||
| Sequence |
MMRTQCLLGLRTFVAFAAKLWSFFIYLLRRQIRTVIQYQTVRYDILPLSPVSRNRLAQVK
RKILVLDLDETLIHSHHDGVLRPTVRPGTPPDFILKVVIDKHPVRFFVHKRPHVDFFLEV VSQWYELVVFTASMEIYGSAVADKLDNSRSILKRRYYRQHCTLELGSYIKDLSVVHSDLS SIVILDNSPGAYRSHPDNAIPIKSWFSDPSDTALLNLLPMLDALRFTADVRSVLSRNLHQ HRLW |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Dullard family
|
|||||
| Subcellular location |
Endoplasmic reticulum membrane
|
|||||
| Function |
Serine/threonine protein phosphatase forming with CNEP1R1 an active phosphatase complex that dephosphorylates and may activate LPIN1 and LPIN2. LPIN1 and LPIN2 are phosphatidate phosphatases that catalyze the conversion of phosphatidic acid to diacylglycerol and control the metabolism of fatty acids at different levels. May indirectly modulate the lipid composition of nuclear and/or endoplasmic reticulum membranes and be required for proper nuclear membrane morphology and/or dynamics. May also indirectly regulate the production of lipid droplets and triacylglycerol. May antagonize BMP signaling.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FBP2 Probe Info |
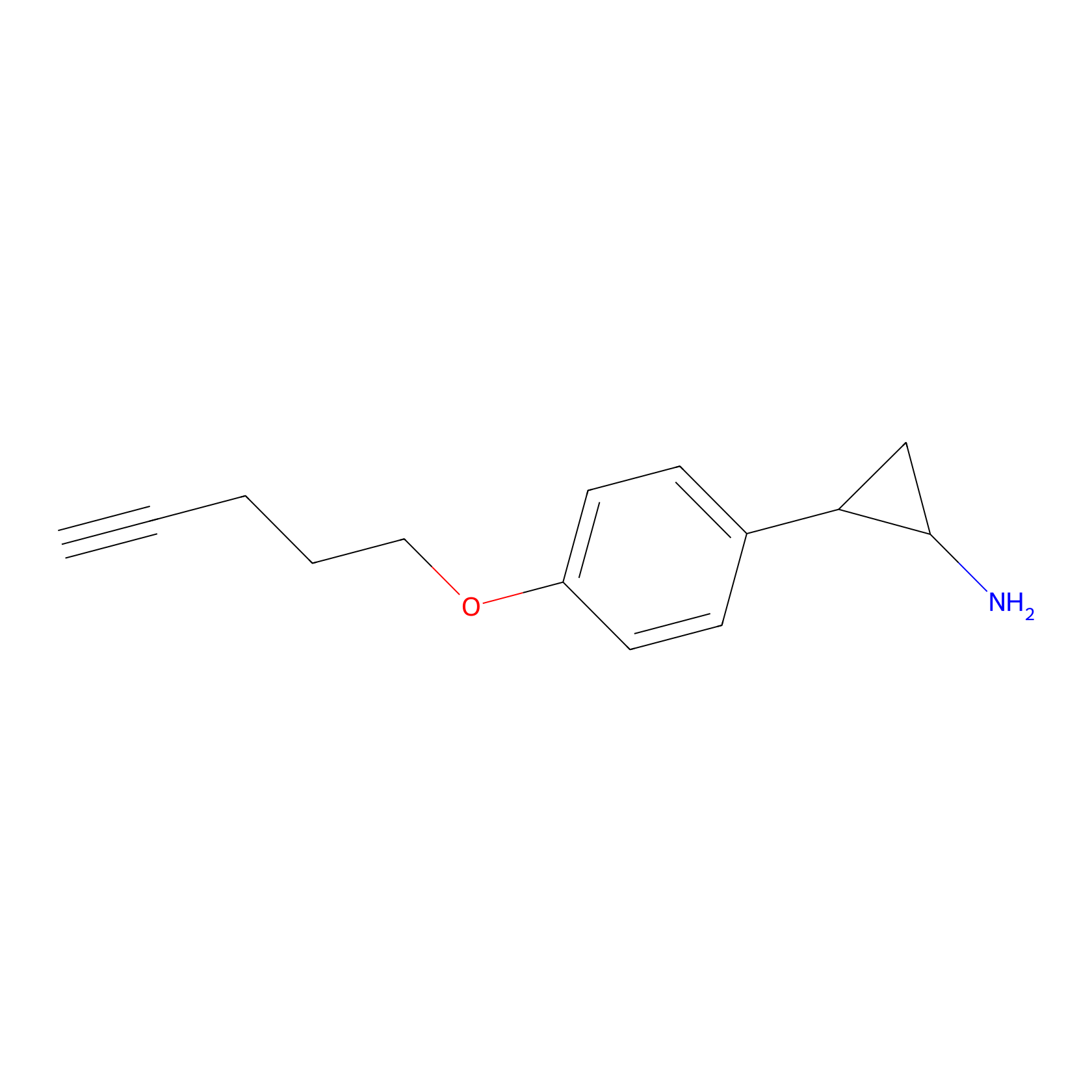 |
3.14 | LDD0317 | [1] | |
|
IPM Probe Info |
 |
N.A. | LDD0241 | [2] | |
|
BTD Probe Info |
 |
C6(2.43) | LDD2100 | [3] | |
|
AHL-Pu-1 Probe Info |
 |
C6(3.69) | LDD0170 | [4] | |
|
IA-alkyne Probe Info |
 |
C6(4.85) | LDD2187 | [5] | |
|
DBIA Probe Info |
 |
C6(0.96) | LDD0580 | [6] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [7] | |
|
YN-1 Probe Info |
 |
N.A. | LDD0446 | [8] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | DM93 | C6(3.69) | LDD0170 | [4] |
| LDCM0026 | 4SU-RNA+native RNA | DM93 | C6(9.51) | LDD0171 | [4] |
| LDCM0215 | AC10 | HEK-293T | C6(0.87) | LDD1508 | [9] |
| LDCM0226 | AC11 | HEK-293T | C6(0.99) | LDD1509 | [9] |
| LDCM0263 | AC143 | HCT 116 | C6(0.96) | LDD0580 | [6] |
| LDCM0264 | AC144 | HCT 116 | C6(0.51) | LDD0581 | [6] |
| LDCM0265 | AC145 | HCT 116 | C6(0.72) | LDD0582 | [6] |
| LDCM0266 | AC146 | HCT 116 | C6(0.85) | LDD0583 | [6] |
| LDCM0267 | AC147 | HCT 116 | C6(0.80) | LDD0584 | [6] |
| LDCM0268 | AC148 | HCT 116 | C6(0.67) | LDD0585 | [6] |
| LDCM0269 | AC149 | HCT 116 | C6(0.73) | LDD0586 | [6] |
| LDCM0271 | AC150 | HCT 116 | C6(0.69) | LDD0588 | [6] |
| LDCM0272 | AC151 | HCT 116 | C6(0.86) | LDD0589 | [6] |
| LDCM0273 | AC152 | HCT 116 | C6(0.87) | LDD0590 | [6] |
| LDCM0274 | AC153 | HCT 116 | C6(0.76) | LDD0591 | [6] |
| LDCM0621 | AC154 | HCT 116 | C6(0.63) | LDD2158 | [6] |
| LDCM0622 | AC155 | HCT 116 | C6(0.83) | LDD2159 | [6] |
| LDCM0623 | AC156 | HCT 116 | C6(0.78) | LDD2160 | [6] |
| LDCM0624 | AC157 | HCT 116 | C6(1.03) | LDD2161 | [6] |
| LDCM0276 | AC17 | HCT 116 | C6(1.07) | LDD0593 | [6] |
| LDCM0277 | AC18 | HCT 116 | C6(0.91) | LDD0594 | [6] |
| LDCM0278 | AC19 | HCT 116 | C6(1.28) | LDD0595 | [6] |
| LDCM0279 | AC2 | HEK-293T | C6(0.91) | LDD1518 | [9] |
| LDCM0280 | AC20 | HCT 116 | C6(1.27) | LDD0597 | [6] |
| LDCM0281 | AC21 | HCT 116 | C6(1.25) | LDD0598 | [6] |
| LDCM0282 | AC22 | HCT 116 | C6(0.82) | LDD0599 | [6] |
| LDCM0283 | AC23 | HCT 116 | C6(1.08) | LDD0600 | [6] |
| LDCM0284 | AC24 | HCT 116 | C6(1.07) | LDD0601 | [6] |
| LDCM0285 | AC25 | HCT 116 | C6(0.71) | LDD0602 | [6] |
| LDCM0286 | AC26 | HCT 116 | C6(1.18) | LDD0603 | [6] |
| LDCM0287 | AC27 | HCT 116 | C6(0.86) | LDD0604 | [6] |
| LDCM0288 | AC28 | HCT 116 | C6(1.19) | LDD0605 | [6] |
| LDCM0289 | AC29 | HCT 116 | C6(1.08) | LDD0606 | [6] |
| LDCM0290 | AC3 | HEK-293T | C6(0.95) | LDD1529 | [9] |
| LDCM0291 | AC30 | HCT 116 | C6(0.98) | LDD0608 | [6] |
| LDCM0292 | AC31 | HCT 116 | C6(0.95) | LDD0609 | [6] |
| LDCM0293 | AC32 | HCT 116 | C6(0.92) | LDD0610 | [6] |
| LDCM0294 | AC33 | HCT 116 | C6(0.92) | LDD0611 | [6] |
| LDCM0295 | AC34 | HCT 116 | C6(0.67) | LDD0612 | [6] |
| LDCM0296 | AC35 | HCT 116 | C6(0.98) | LDD0613 | [6] |
| LDCM0297 | AC36 | HCT 116 | C6(1.05) | LDD0614 | [6] |
| LDCM0298 | AC37 | HCT 116 | C6(1.35) | LDD0615 | [6] |
| LDCM0299 | AC38 | HCT 116 | C6(0.90) | LDD0616 | [6] |
| LDCM0300 | AC39 | HCT 116 | C6(1.07) | LDD0617 | [6] |
| LDCM0302 | AC40 | HCT 116 | C6(1.04) | LDD0619 | [6] |
| LDCM0303 | AC41 | HCT 116 | C6(1.34) | LDD0620 | [6] |
| LDCM0304 | AC42 | HCT 116 | C6(1.23) | LDD0621 | [6] |
| LDCM0305 | AC43 | HCT 116 | C6(1.21) | LDD0622 | [6] |
| LDCM0306 | AC44 | HCT 116 | C6(1.10) | LDD0623 | [6] |
| LDCM0307 | AC45 | HCT 116 | C6(1.48) | LDD0624 | [6] |
| LDCM0310 | AC48 | HEK-293T | C6(1.02) | LDD1549 | [9] |
| LDCM0313 | AC50 | HEK-293T | C6(0.97) | LDD1552 | [9] |
| LDCM0314 | AC51 | HEK-293T | C6(0.95) | LDD1553 | [9] |
| LDCM0319 | AC56 | HEK-293T | C6(1.03) | LDD1558 | [9] |
| LDCM0321 | AC58 | HEK-293T | C6(1.02) | LDD1560 | [9] |
| LDCM0322 | AC59 | HEK-293T | C6(0.97) | LDD1561 | [9] |
| LDCM0328 | AC64 | HEK-293T | C6(0.92) | LDD1567 | [9] |
| LDCM0345 | AC8 | HEK-293T | C6(1.01) | LDD1569 | [9] |
| LDCM0275 | AKOS034007705 | HEK-293T | C6(1.04) | LDD1514 | [9] |
| LDCM0367 | CL1 | HCT 116 | C6(0.73) | LDD0684 | [6] |
| LDCM0368 | CL10 | HCT 116 | C6(0.55) | LDD0685 | [6] |
| LDCM0370 | CL101 | HEK-293T | C6(1.10) | LDD1574 | [9] |
| LDCM0374 | CL105 | HCT 116 | C6(1.69) | LDD0691 | [6] |
| LDCM0375 | CL106 | HCT 116 | C6(1.12) | LDD0692 | [6] |
| LDCM0376 | CL107 | HCT 116 | C6(1.30) | LDD0693 | [6] |
| LDCM0377 | CL108 | HCT 116 | C6(1.09) | LDD0694 | [6] |
| LDCM0378 | CL109 | HCT 116 | C6(0.99) | LDD0695 | [6] |
| LDCM0379 | CL11 | HCT 116 | C6(0.74) | LDD0696 | [6] |
| LDCM0380 | CL110 | HCT 116 | C6(1.01) | LDD0697 | [6] |
| LDCM0381 | CL111 | HCT 116 | C6(1.22) | LDD0698 | [6] |
| LDCM0382 | CL112 | HCT 116 | C6(0.92) | LDD0699 | [6] |
| LDCM0383 | CL113 | HCT 116 | C6(0.92) | LDD0700 | [6] |
| LDCM0384 | CL114 | HCT 116 | C6(0.98) | LDD0701 | [6] |
| LDCM0385 | CL115 | HCT 116 | C6(0.89) | LDD0702 | [6] |
| LDCM0386 | CL116 | HCT 116 | C6(0.82) | LDD0703 | [6] |
| LDCM0387 | CL117 | HCT 116 | C6(1.01) | LDD0704 | [6] |
| LDCM0388 | CL118 | HCT 116 | C6(1.47) | LDD0705 | [6] |
| LDCM0389 | CL119 | HCT 116 | C6(1.07) | LDD0706 | [6] |
| LDCM0390 | CL12 | HCT 116 | C6(0.60) | LDD0707 | [6] |
| LDCM0391 | CL120 | HCT 116 | C6(1.25) | LDD0708 | [6] |
| LDCM0392 | CL121 | HEK-293T | C6(1.09) | LDD1596 | [9] |
| LDCM0396 | CL125 | HEK-293T | C6(1.04) | LDD1600 | [9] |
| LDCM0400 | CL13 | HCT 116 | C6(0.67) | LDD0717 | [6] |
| LDCM0401 | CL14 | HCT 116 | C6(0.73) | LDD0718 | [6] |
| LDCM0402 | CL15 | HCT 116 | C6(0.96) | LDD0719 | [6] |
| LDCM0405 | CL18 | HEK-293T | C6(1.10) | LDD1609 | [9] |
| LDCM0406 | CL19 | HEK-293T | C6(1.25) | LDD1610 | [9] |
| LDCM0407 | CL2 | HCT 116 | C6(0.58) | LDD0724 | [6] |
| LDCM0412 | CL24 | HEK-293T | C6(1.52) | LDD1616 | [9] |
| LDCM0413 | CL25 | HEK-293T | C6(1.51) | LDD1617 | [9] |
| LDCM0418 | CL3 | HCT 116 | C6(0.67) | LDD0735 | [6] |
| LDCM0419 | CL30 | HEK-293T | C6(0.91) | LDD1623 | [9] |
| LDCM0420 | CL31 | HEK-293T | C6(1.27) | LDD1624 | [9] |
| LDCM0425 | CL36 | HEK-293T | C6(1.48) | LDD1629 | [9] |
| LDCM0426 | CL37 | HEK-293T | C6(1.22) | LDD1630 | [9] |
| LDCM0429 | CL4 | HCT 116 | C6(1.29) | LDD0746 | [6] |
| LDCM0432 | CL42 | HEK-293T | C6(0.97) | LDD1636 | [9] |
| LDCM0433 | CL43 | HEK-293T | C6(1.05) | LDD1637 | [9] |
| LDCM0438 | CL48 | HEK-293T | C6(1.21) | LDD1642 | [9] |
| LDCM0439 | CL49 | HEK-293T | C6(1.20) | LDD1643 | [9] |
| LDCM0440 | CL5 | HCT 116 | C6(0.56) | LDD0757 | [6] |
| LDCM0445 | CL54 | HEK-293T | C6(1.55) | LDD1648 | [9] |
| LDCM0446 | CL55 | HEK-293T | C6(1.16) | LDD1649 | [9] |
| LDCM0451 | CL6 | HCT 116 | C6(0.77) | LDD0768 | [6] |
| LDCM0452 | CL60 | HEK-293T | C6(1.29) | LDD1655 | [9] |
| LDCM0453 | CL61 | HCT 116 | C6(1.31) | LDD0770 | [6] |
| LDCM0454 | CL62 | HCT 116 | C6(1.11) | LDD0771 | [6] |
| LDCM0455 | CL63 | HCT 116 | C6(1.34) | LDD0772 | [6] |
| LDCM0456 | CL64 | HCT 116 | C6(1.19) | LDD0773 | [6] |
| LDCM0457 | CL65 | HCT 116 | C6(1.06) | LDD0774 | [6] |
| LDCM0458 | CL66 | HCT 116 | C6(1.09) | LDD0775 | [6] |
| LDCM0459 | CL67 | HCT 116 | C6(1.02) | LDD0776 | [6] |
| LDCM0460 | CL68 | HCT 116 | C6(0.96) | LDD0777 | [6] |
| LDCM0461 | CL69 | HCT 116 | C6(1.39) | LDD0778 | [6] |
| LDCM0462 | CL7 | HCT 116 | C6(0.82) | LDD0779 | [6] |
| LDCM0463 | CL70 | HCT 116 | C6(0.97) | LDD0780 | [6] |
| LDCM0464 | CL71 | HCT 116 | C6(1.00) | LDD0781 | [6] |
| LDCM0465 | CL72 | HCT 116 | C6(1.05) | LDD0782 | [6] |
| LDCM0466 | CL73 | HCT 116 | C6(1.25) | LDD0783 | [6] |
| LDCM0467 | CL74 | HCT 116 | C6(1.14) | LDD0784 | [6] |
| LDCM0469 | CL76 | HCT 116 | C6(0.92) | LDD0786 | [6] |
| LDCM0470 | CL77 | HCT 116 | C6(0.79) | LDD0787 | [6] |
| LDCM0471 | CL78 | HCT 116 | C6(0.85) | LDD0788 | [6] |
| LDCM0472 | CL79 | HCT 116 | C6(0.77) | LDD0789 | [6] |
| LDCM0473 | CL8 | HCT 116 | C6(0.68) | LDD0790 | [6] |
| LDCM0474 | CL80 | HCT 116 | C6(0.89) | LDD0791 | [6] |
| LDCM0475 | CL81 | HCT 116 | C6(0.86) | LDD0792 | [6] |
| LDCM0476 | CL82 | HCT 116 | C6(0.84) | LDD0793 | [6] |
| LDCM0477 | CL83 | HCT 116 | C6(0.98) | LDD0794 | [6] |
| LDCM0478 | CL84 | HCT 116 | C6(0.87) | LDD0795 | [6] |
| LDCM0479 | CL85 | HCT 116 | C6(0.91) | LDD0796 | [6] |
| LDCM0480 | CL86 | HCT 116 | C6(0.92) | LDD0797 | [6] |
| LDCM0481 | CL87 | HCT 116 | C6(0.71) | LDD0798 | [6] |
| LDCM0482 | CL88 | HCT 116 | C6(0.73) | LDD0799 | [6] |
| LDCM0483 | CL89 | HCT 116 | C6(0.98) | LDD0800 | [6] |
| LDCM0484 | CL9 | HCT 116 | C6(0.69) | LDD0801 | [6] |
| LDCM0485 | CL90 | HCT 116 | C6(0.87) | LDD0802 | [6] |
| LDCM0486 | CL91 | HEK-293T | C6(1.08) | LDD1689 | [9] |
| LDCM0491 | CL96 | HEK-293T | C6(1.42) | LDD1694 | [9] |
| LDCM0492 | CL97 | HEK-293T | C6(1.53) | LDD1695 | [9] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C6(8.64) | LDD1702 | [3] |
| LDCM0625 | F8 | Ramos | C6(4.85) | LDD2187 | [5] |
| LDCM0573 | Fragment11 | Ramos | C6(0.56) | LDD2190 | [5] |
| LDCM0575 | Fragment13 | Ramos | C6(3.87) | LDD2192 | [5] |
| LDCM0582 | Fragment23 | Ramos | C6(1.81) | LDD2196 | [5] |
| LDCM0578 | Fragment27 | Ramos | C6(1.07) | LDD2197 | [5] |
| LDCM0586 | Fragment28 | Ramos | C6(1.47) | LDD2198 | [5] |
| LDCM0468 | Fragment33 | HCT 116 | C6(0.93) | LDD0785 | [6] |
| LDCM0614 | Fragment56 | Ramos | C6(1.49) | LDD2205 | [5] |
| LDCM0022 | KB02 | GB-2 | C6(1.62) | LDD2335 | [10] |
| LDCM0023 | KB03 | DM93 | C6(1.89) | LDD2889 | [10] |
| LDCM0024 | KB05 | SKMEL24 | C6(1.60) | LDD3323 | [10] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C6(2.43) | LDD2100 | [3] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C6(1.23) | LDD2105 | [3] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C6(0.64) | LDD2137 | [3] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C6(0.61) | LDD2140 | [3] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C6(0.90) | LDD2141 | [3] |
| LDCM0551 | Nucleophilic fragment 5b | MDA-MB-231 | C6(5.47) | LDD2145 | [3] |
The Interaction Atlas With This Target
References
