Details of the Target
General Information of Target
| Target ID | LDTP15778 | |||||
|---|---|---|---|---|---|---|
| Target Name | Protein PRRC1 (PRRC1) | |||||
| Gene Name | PRRC1 | |||||
| Gene ID | 133619 | |||||
| Synonyms |
Protein PRRC1; Proline-rich and coiled-coil-containing protein 1 |
|||||
| 3D Structure | ||||||
| Sequence |
MSGAGVAAGTRPPSSPTPGSRRRRQRPSVGVQSLRPQSPQLRQSDPQKRNLDLEKSLQFL
QQQHSEMLAKLHEEIEHLKRENKDLRYKLIMNQTSQKKDGPSGNHLSRASAPLGARWVCI NGVWVEPGGPSPARLKEGSSRTHRPGGKHGRLAGGSADTVRSPADSLSTSSFQSVKSISN SGKARPQPGSFNKQDSKADVPQKADLEEEPLLHNSKLDKVPGVQGQARKEKAEASNAGAA CMGNSQHQGRQMGAAAHPPMILPLPLRKPTTLRQCEVLIRELWNTNLLQTQELQHLKSLL EGSQRPQAVPEEASFPRDQEATHFPKVSTKSLSKKCLLLSPPVAERAILPALKQTPKNNF AERQKRLQAMQKRRLHRSVL |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
PRRC1 family
|
|||||
| Subcellular location |
Golgi apparatus
|
|||||
| Function | May act as a regulator of the protein kinase A (PKA) activity during embryonic development. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
AZ-9 Probe Info |
 |
4.15 | LDD0393 | [1] | |
|
STPyne Probe Info |
 |
K218(6.67); K227(2.66); K232(6.39); K234(4.02) | LDD0277 | [2] | |
|
HHS-475 Probe Info |
 |
Y251(0.80) | LDD0264 | [3] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0221 | [4] | |
|
DBIA Probe Info |
 |
C348(0.94) | LDD1510 | [5] | |
|
ATP probe Probe Info |
 |
K266(0.00); K269(0.00) | LDD0199 | [6] | |
|
m-APA Probe Info |
 |
N.A. | LDD2231 | [7] | |
|
AOyne Probe Info |
 |
10.20 | LDD0443 | [8] | |
|
HHS-465 Probe Info |
 |
Y251(0.00); K253(0.00) | LDD2240 | [9] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C112 Probe Info |
 |
21.71 | LDD1799 | [10] | |
|
C232 Probe Info |
 |
41.36 | LDD1905 | [10] | |
|
C235 Probe Info |
 |
21.11 | LDD1908 | [10] | |
|
C284 Probe Info |
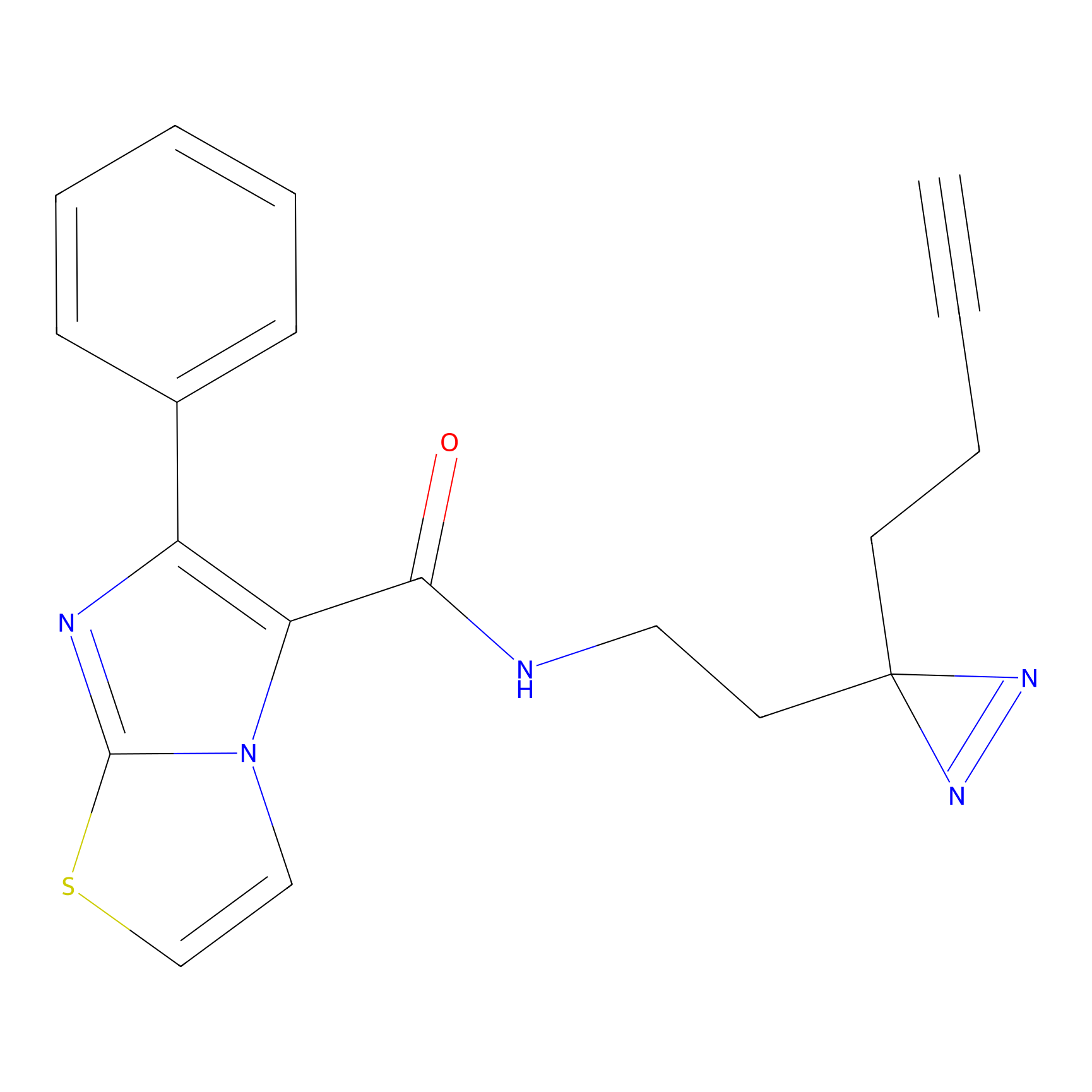 |
22.32 | LDD1954 | [10] | |
|
C362 Probe Info |
 |
36.76 | LDD2023 | [10] | |
|
C382 Probe Info |
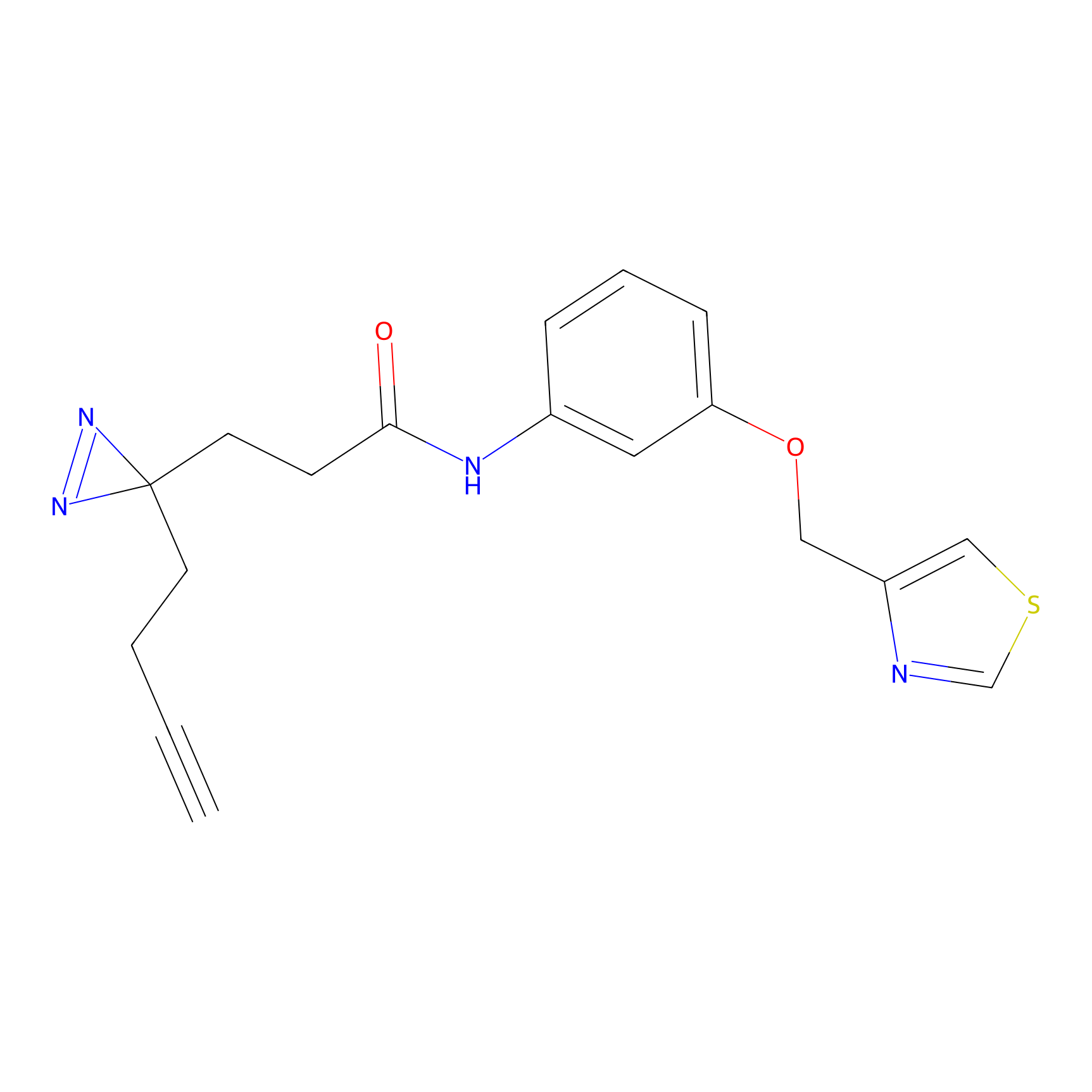 |
9.45 | LDD2041 | [10] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0237 | AC12 | HEK-293T | C348(0.94) | LDD1510 | [5] |
| LDCM0259 | AC14 | HEK-293T | C348(1.11) | LDD1512 | [5] |
| LDCM0280 | AC20 | HEK-293T | C348(0.85) | LDD1519 | [5] |
| LDCM0282 | AC22 | HEK-293T | C348(1.10) | LDD1521 | [5] |
| LDCM0288 | AC28 | HEK-293T | C348(1.09) | LDD1527 | [5] |
| LDCM0291 | AC30 | HEK-293T | C348(1.48) | LDD1530 | [5] |
| LDCM0297 | AC36 | HEK-293T | C348(0.97) | LDD1536 | [5] |
| LDCM0299 | AC38 | HEK-293T | C348(1.08) | LDD1538 | [5] |
| LDCM0301 | AC4 | HEK-293T | C348(1.11) | LDD1540 | [5] |
| LDCM0306 | AC44 | HEK-293T | C348(0.92) | LDD1545 | [5] |
| LDCM0308 | AC46 | HEK-293T | C348(1.27) | LDD1547 | [5] |
| LDCM0315 | AC52 | HEK-293T | C348(1.03) | LDD1554 | [5] |
| LDCM0317 | AC54 | HEK-293T | C348(1.13) | LDD1556 | [5] |
| LDCM0323 | AC6 | HEK-293T | C348(1.04) | LDD1562 | [5] |
| LDCM0324 | AC60 | HEK-293T | C348(1.10) | LDD1563 | [5] |
| LDCM0326 | AC62 | HEK-293T | C348(1.04) | LDD1565 | [5] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [4] |
| LDCM0368 | CL10 | HEK-293T | C348(1.35) | LDD1572 | [5] |
| LDCM0371 | CL102 | HEK-293T | C348(0.90) | LDD1575 | [5] |
| LDCM0375 | CL106 | HEK-293T | C348(0.92) | LDD1579 | [5] |
| LDCM0380 | CL110 | HEK-293T | C348(1.05) | LDD1584 | [5] |
| LDCM0384 | CL114 | HEK-293T | C348(1.30) | LDD1588 | [5] |
| LDCM0388 | CL118 | HEK-293T | C348(0.96) | LDD1592 | [5] |
| LDCM0393 | CL122 | HEK-293T | C348(1.15) | LDD1597 | [5] |
| LDCM0397 | CL126 | HEK-293T | C348(1.15) | LDD1601 | [5] |
| LDCM0401 | CL14 | HEK-293T | C348(1.03) | LDD1605 | [5] |
| LDCM0407 | CL2 | HEK-293T | C348(0.97) | LDD1611 | [5] |
| LDCM0408 | CL20 | HEK-293T | C348(1.20) | LDD1612 | [5] |
| LDCM0410 | CL22 | HEK-293T | C348(1.28) | LDD1614 | [5] |
| LDCM0414 | CL26 | HEK-293T | C348(0.94) | LDD1618 | [5] |
| LDCM0421 | CL32 | HEK-293T | C348(1.23) | LDD1625 | [5] |
| LDCM0423 | CL34 | HEK-293T | C348(1.16) | LDD1627 | [5] |
| LDCM0434 | CL44 | HEK-293T | C348(1.16) | LDD1638 | [5] |
| LDCM0436 | CL46 | HEK-293T | C348(1.61) | LDD1640 | [5] |
| LDCM0441 | CL50 | HEK-293T | C348(1.17) | LDD1645 | [5] |
| LDCM0447 | CL56 | HEK-293T | C348(0.94) | LDD1650 | [5] |
| LDCM0449 | CL58 | HEK-293T | C348(1.30) | LDD1652 | [5] |
| LDCM0454 | CL62 | HEK-293T | C348(1.05) | LDD1657 | [5] |
| LDCM0460 | CL68 | HEK-293T | C348(1.06) | LDD1663 | [5] |
| LDCM0463 | CL70 | HEK-293T | C348(1.31) | LDD1666 | [5] |
| LDCM0467 | CL74 | HEK-293T | C348(1.53) | LDD1670 | [5] |
| LDCM0473 | CL8 | HEK-293T | C348(1.76) | LDD1676 | [5] |
| LDCM0474 | CL80 | HEK-293T | C348(1.12) | LDD1677 | [5] |
| LDCM0476 | CL82 | HEK-293T | C348(1.15) | LDD1679 | [5] |
| LDCM0480 | CL86 | HEK-293T | C348(1.36) | LDD1683 | [5] |
| LDCM0487 | CL92 | HEK-293T | C348(1.05) | LDD1690 | [5] |
| LDCM0489 | CL94 | HEK-293T | C348(1.19) | LDD1692 | [5] |
| LDCM0493 | CL98 | HEK-293T | C348(1.14) | LDD1696 | [5] |
| LDCM0427 | Fragment51 | HEK-293T | C348(0.85) | LDD1631 | [5] |
| LDCM0116 | HHS-0101 | DM93 | Y251(0.80) | LDD0264 | [3] |
| LDCM0117 | HHS-0201 | DM93 | Y251(0.85) | LDD0265 | [3] |
| LDCM0118 | HHS-0301 | DM93 | Y251(0.86) | LDD0266 | [3] |
| LDCM0119 | HHS-0401 | DM93 | Y251(0.81) | LDD0267 | [3] |
| LDCM0120 | HHS-0701 | DM93 | Y251(1.46) | LDD0268 | [3] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [4] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (PIN1) | . | Q13526 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Huntingtin (HTT) | Huntingtin family | P42858 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Protein transport protein Sec23B (SEC23B) | SEC23/SEC24 family | Q15437 | |||
References
