Details of the Target
General Information of Target
| Target ID | LDTP14421 | |||||
|---|---|---|---|---|---|---|
| Target Name | Ladinin-1 (LAD1) | |||||
| Gene Name | LAD1 | |||||
| Gene ID | 3898 | |||||
| Synonyms |
LAD; Ladinin-1; Lad-1; Linear IgA disease antigen; LADA |
|||||
| 3D Structure | ||||||
| Sequence |
MPKNKKRNTPHRGSSAGGGGSGAAAATAATAGGQHRNVQPFSDEDASIETMSHCSGYSDP
SSFAEDGPEVLDEEGTQEDLEYKLKGLIDLTLDKSAKTRQAALEGIKNALASKMLYEFIL ERRMTLTDSIERCLKKGKSDEQRAAAALASVLCIQLGPGIESEEILKTLGPILKKIICDG SASMQARQTCATCFGVCCFIATDDITELYSTLECLENIFTKSYLKEKDTTVICSTPNTVL HISSLLAWTLLLTICPINEVKKKLEMHFHKLPSLLSCDDVNMRIAAGESLALLFELARGI ESDFFYEDMESLTQMLRALATDGNKHRAKVDKRKQRSVFRDVLRAVEERDFPTETIKFGP ERMYIDCWVKKHTYDTFKEVLGSGMQYHLQSNEFLRNVFELGPPVMLDAATLKTMKISRF ERHLYNSAAFKARTKARSKCRDKRADVGEFF |
|||||
| Target Bioclass |
Other
|
|||||
| Subcellular location |
Secreted, extracellular space, extracellular matrix, basement membrane
|
|||||
| Function | Anchoring filament protein which is a component of the basement membrane zone. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TG42 Probe Info |
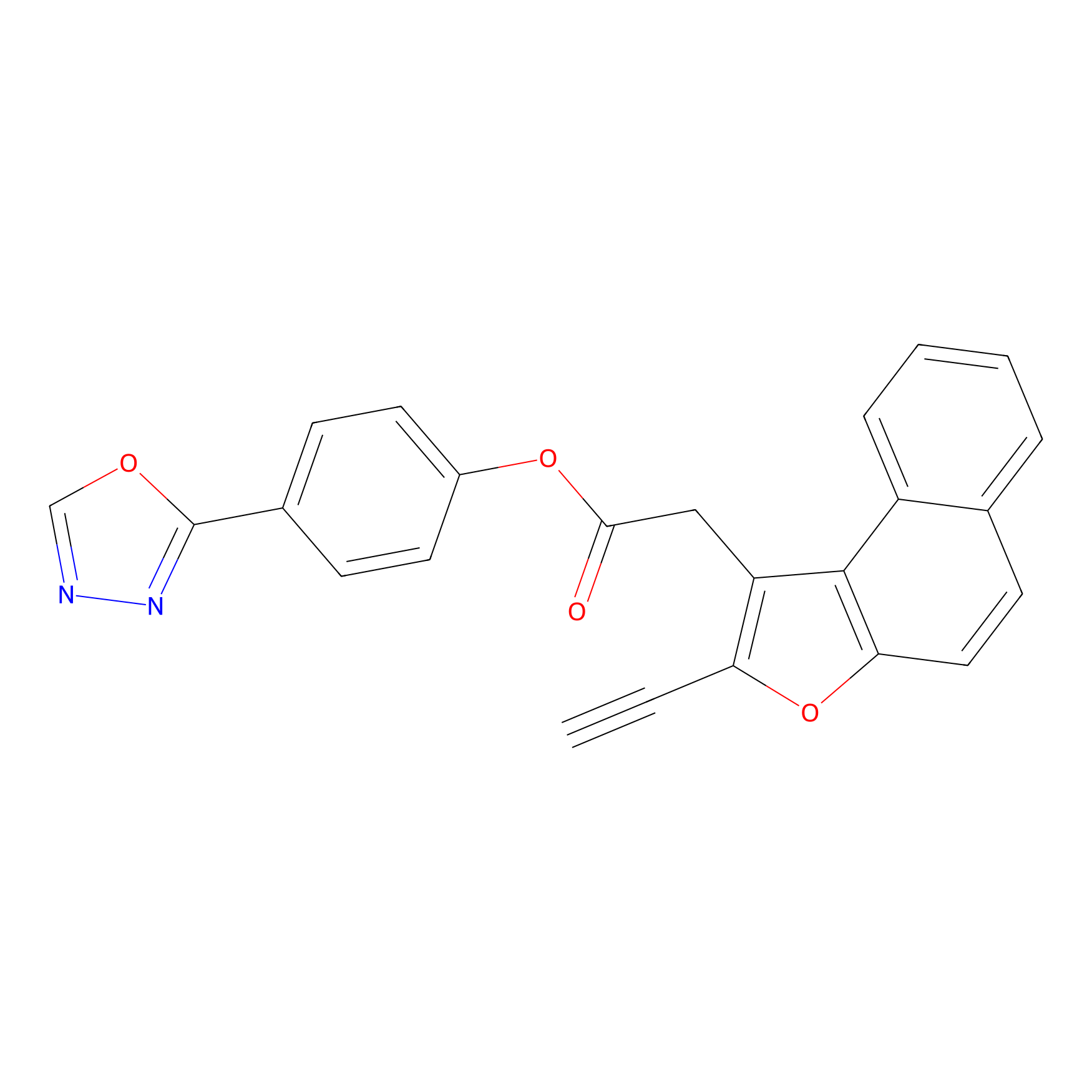 |
5.82 | LDD0326 | [1] | |
|
C-Sul Probe Info |
 |
31.37 | LDD0066 | [2] | |
|
AZ-9 Probe Info |
 |
E116(1.37) | LDD2209 | [3] | |
|
IPM Probe Info |
 |
N.A. | LDD0241 | [4] | |
|
Probe 1 Probe Info |
 |
Y362(83.92) | LDD3495 | [5] | |
|
DBIA Probe Info |
 |
C428(0.96) | LDD0531 | [6] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0149 | [7] | |
|
PF-06672131 Probe Info |
 |
N.A. | LDD0017 | [8] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [4] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [9] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HCT 116 | C428(0.96) | LDD0531 | [6] |
| LDCM0216 | AC100 | HCT 116 | C428(0.93) | LDD0533 | [6] |
| LDCM0217 | AC101 | HCT 116 | C428(0.92) | LDD0534 | [6] |
| LDCM0218 | AC102 | HCT 116 | C428(0.84) | LDD0535 | [6] |
| LDCM0219 | AC103 | HCT 116 | C428(1.12) | LDD0536 | [6] |
| LDCM0220 | AC104 | HCT 116 | C428(0.95) | LDD0537 | [6] |
| LDCM0221 | AC105 | HCT 116 | C428(0.97) | LDD0538 | [6] |
| LDCM0222 | AC106 | HCT 116 | C428(0.95) | LDD0539 | [6] |
| LDCM0223 | AC107 | HCT 116 | C428(0.95) | LDD0540 | [6] |
| LDCM0224 | AC108 | HCT 116 | C428(0.83) | LDD0541 | [6] |
| LDCM0225 | AC109 | HCT 116 | C428(0.81) | LDD0542 | [6] |
| LDCM0227 | AC110 | HCT 116 | C428(0.81) | LDD0544 | [6] |
| LDCM0228 | AC111 | HCT 116 | C428(1.01) | LDD0545 | [6] |
| LDCM0229 | AC112 | HCT 116 | C428(1.13) | LDD0546 | [6] |
| LDCM0246 | AC128 | HCT 116 | C428(1.10) | LDD0563 | [6] |
| LDCM0247 | AC129 | HCT 116 | C428(0.93) | LDD0564 | [6] |
| LDCM0249 | AC130 | HCT 116 | C428(1.03) | LDD0566 | [6] |
| LDCM0250 | AC131 | HCT 116 | C428(1.32) | LDD0567 | [6] |
| LDCM0251 | AC132 | HCT 116 | C428(0.72) | LDD0568 | [6] |
| LDCM0252 | AC133 | HCT 116 | C428(0.87) | LDD0569 | [6] |
| LDCM0253 | AC134 | HCT 116 | C428(1.04) | LDD0570 | [6] |
| LDCM0254 | AC135 | HCT 116 | C428(0.85) | LDD0571 | [6] |
| LDCM0255 | AC136 | HCT 116 | C428(0.93) | LDD0572 | [6] |
| LDCM0256 | AC137 | HCT 116 | C428(0.73) | LDD0573 | [6] |
| LDCM0257 | AC138 | HCT 116 | C428(0.72) | LDD0574 | [6] |
| LDCM0258 | AC139 | HCT 116 | C428(0.86) | LDD0575 | [6] |
| LDCM0260 | AC140 | HCT 116 | C428(0.89) | LDD0577 | [6] |
| LDCM0261 | AC141 | HCT 116 | C428(0.87) | LDD0578 | [6] |
| LDCM0262 | AC142 | HCT 116 | C428(0.74) | LDD0579 | [6] |
| LDCM0263 | AC143 | HCT 116 | C428(1.14) | LDD0580 | [6] |
| LDCM0264 | AC144 | HCT 116 | C428(0.97) | LDD0581 | [6] |
| LDCM0265 | AC145 | HCT 116 | C428(1.04) | LDD0582 | [6] |
| LDCM0266 | AC146 | HCT 116 | C428(1.15) | LDD0583 | [6] |
| LDCM0267 | AC147 | HCT 116 | C428(0.76) | LDD0584 | [6] |
| LDCM0268 | AC148 | HCT 116 | C428(0.74) | LDD0585 | [6] |
| LDCM0269 | AC149 | HCT 116 | C428(1.16) | LDD0586 | [6] |
| LDCM0271 | AC150 | HCT 116 | C428(1.01) | LDD0588 | [6] |
| LDCM0272 | AC151 | HCT 116 | C428(1.30) | LDD0589 | [6] |
| LDCM0273 | AC152 | HCT 116 | C428(1.08) | LDD0590 | [6] |
| LDCM0274 | AC153 | HCT 116 | C428(0.49) | LDD0591 | [6] |
| LDCM0621 | AC154 | HCT 116 | C428(1.24) | LDD2158 | [6] |
| LDCM0622 | AC155 | HCT 116 | C428(0.92) | LDD2159 | [6] |
| LDCM0623 | AC156 | HCT 116 | C428(1.05) | LDD2160 | [6] |
| LDCM0624 | AC157 | HCT 116 | C428(1.06) | LDD2161 | [6] |
| LDCM0279 | AC2 | HCT 116 | C428(1.12) | LDD0596 | [6] |
| LDCM0290 | AC3 | HCT 116 | C428(1.00) | LDD0607 | [6] |
| LDCM0301 | AC4 | HCT 116 | C428(0.94) | LDD0618 | [6] |
| LDCM0312 | AC5 | HCT 116 | C428(0.84) | LDD0629 | [6] |
| LDCM0320 | AC57 | HCT 116 | C428(1.05) | LDD0637 | [6] |
| LDCM0321 | AC58 | HCT 116 | C428(1.06) | LDD0638 | [6] |
| LDCM0322 | AC59 | HCT 116 | C428(1.00) | LDD0639 | [6] |
| LDCM0324 | AC60 | HCT 116 | C428(1.18) | LDD0641 | [6] |
| LDCM0325 | AC61 | HCT 116 | C428(0.87) | LDD0642 | [6] |
| LDCM0326 | AC62 | HCT 116 | C428(1.04) | LDD0643 | [6] |
| LDCM0327 | AC63 | HCT 116 | C428(1.30) | LDD0644 | [6] |
| LDCM0328 | AC64 | HCT 116 | C428(1.13) | LDD0645 | [6] |
| LDCM0329 | AC65 | HCT 116 | C428(1.10) | LDD0646 | [6] |
| LDCM0330 | AC66 | HCT 116 | C428(1.08) | LDD0647 | [6] |
| LDCM0331 | AC67 | HCT 116 | C428(1.07) | LDD0648 | [6] |
| LDCM0365 | AC98 | HCT 116 | C428(1.32) | LDD0682 | [6] |
| LDCM0366 | AC99 | HCT 116 | C428(0.87) | LDD0683 | [6] |
| LDCM0020 | ARS-1620 | HCC44 | C428(1.17) | LDD2171 | [6] |
| LDCM0632 | CL-Sc | Hep-G2 | C428(2.03) | LDD2227 | [9] |
| LDCM0369 | CL100 | HCT 116 | C428(1.06) | LDD0686 | [6] |
| LDCM0396 | CL125 | HCT 116 | C428(1.25) | LDD0713 | [6] |
| LDCM0397 | CL126 | HCT 116 | C428(1.32) | LDD0714 | [6] |
| LDCM0398 | CL127 | HCT 116 | C428(1.11) | LDD0715 | [6] |
| LDCM0399 | CL128 | HCT 116 | C428(0.97) | LDD0716 | [6] |
| LDCM0436 | CL46 | HCT 116 | C428(1.38) | LDD0753 | [6] |
| LDCM0437 | CL47 | HCT 116 | C428(1.32) | LDD0754 | [6] |
| LDCM0438 | CL48 | HCT 116 | C428(0.97) | LDD0755 | [6] |
| LDCM0439 | CL49 | HCT 116 | C428(1.18) | LDD0756 | [6] |
| LDCM0441 | CL50 | HCT 116 | C428(1.11) | LDD0758 | [6] |
| LDCM0442 | CL51 | HCT 116 | C428(0.91) | LDD0759 | [6] |
| LDCM0443 | CL52 | HCT 116 | C428(0.91) | LDD0760 | [6] |
| LDCM0444 | CL53 | HCT 116 | C428(0.97) | LDD0761 | [6] |
| LDCM0445 | CL54 | HCT 116 | C428(1.27) | LDD0762 | [6] |
| LDCM0446 | CL55 | HCT 116 | C428(1.14) | LDD0763 | [6] |
| LDCM0447 | CL56 | HCT 116 | C428(1.54) | LDD0764 | [6] |
| LDCM0448 | CL57 | HCT 116 | C428(1.17) | LDD0765 | [6] |
| LDCM0449 | CL58 | HCT 116 | C428(1.03) | LDD0766 | [6] |
| LDCM0450 | CL59 | HCT 116 | C428(1.18) | LDD0767 | [6] |
| LDCM0452 | CL60 | HCT 116 | C428(1.89) | LDD0769 | [6] |
| LDCM0486 | CL91 | HCT 116 | C428(1.08) | LDD0803 | [6] |
| LDCM0487 | CL92 | HCT 116 | C428(1.01) | LDD0804 | [6] |
| LDCM0488 | CL93 | HCT 116 | C428(1.13) | LDD0805 | [6] |
| LDCM0489 | CL94 | HCT 116 | C428(1.31) | LDD0806 | [6] |
| LDCM0490 | CL95 | HCT 116 | C428(0.94) | LDD0807 | [6] |
| LDCM0491 | CL96 | HCT 116 | C428(1.08) | LDD0808 | [6] |
| LDCM0492 | CL97 | HCT 116 | C428(1.01) | LDD0809 | [6] |
| LDCM0493 | CL98 | HCT 116 | C428(1.00) | LDD0810 | [6] |
| LDCM0494 | CL99 | HCT 116 | C428(1.11) | LDD0811 | [6] |
| LDCM0021 | THZ1 | HCT 116 | C428(1.17) | LDD2173 | [6] |
| LDCM0111 | W14 | Hep-G2 | C428(23.04) | LDD0238 | [4] |
| LDCM0113 | W17 | Hep-G2 | C428(12.29) | LDD0240 | [4] |
References
