Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
OPA-S-S-alkyne Probe Info |
 |
K559(2.92); K574(5.16) | LDD3494 | [1] | |
|
Probe 1 Probe Info |
 |
Y16(43.38); Y87(8.59); Y555(9.75) | LDD3495 | [2] | |
|
BTD Probe Info |
 |
C62(1.95) | LDD1700 | [3] | |
|
HHS-475 Probe Info |
 |
Y87(0.96); Y38(1.20); Y16(1.27) | LDD0264 | [4] | |
|
HHS-465 Probe Info |
 |
Y16(10.00); Y38(7.45); Y529(10.00); Y70(5.83) | LDD2237 | [5] | |
|
DBIA Probe Info |
 |
C62(0.76) | LDD0531 | [6] | |
|
5E-2FA Probe Info |
 |
H56(0.00); H39(0.00); H78(0.00) | LDD2235 | [7] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [8] | |
|
m-APA Probe Info |
 |
H106(0.00); H56(0.00) | LDD2231 | [7] | |
|
IA-alkyne Probe Info |
 |
C62(0.00); C7(0.00) | LDD0165 | [9] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [10] | |
|
JW-RF-010 Probe Info |
 |
C62(0.00); C7(0.00) | LDD0026 | [11] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [12] | |
|
NHS Probe Info |
 |
K574(0.00); K559(0.00) | LDD0010 | [12] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [13] | |
|
STPyne Probe Info |
 |
K574(0.00); K559(0.00) | LDD0009 | [12] | |
|
YN-1 Probe Info |
 |
N.A. | LDD0446 | [14] | |
|
Ox-W18 Probe Info |
 |
W23(0.00); W258(0.00) | LDD2175 | [15] | |
|
1c-yne Probe Info |
 |
K427(0.00); K94(0.00) | LDD0228 | [16] | |
|
1d-yne Probe Info |
 |
K427(0.00); K461(0.00) | LDD0357 | [16] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [17] | |
|
NAIA_5 Probe Info |
 |
C62(0.00); C7(0.00) | LDD2223 | [18] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C187 Probe Info |
 |
18.51 | LDD1865 | [19] | |
|
C191 Probe Info |
 |
10.85 | LDD1868 | [19] | |
|
C193 Probe Info |
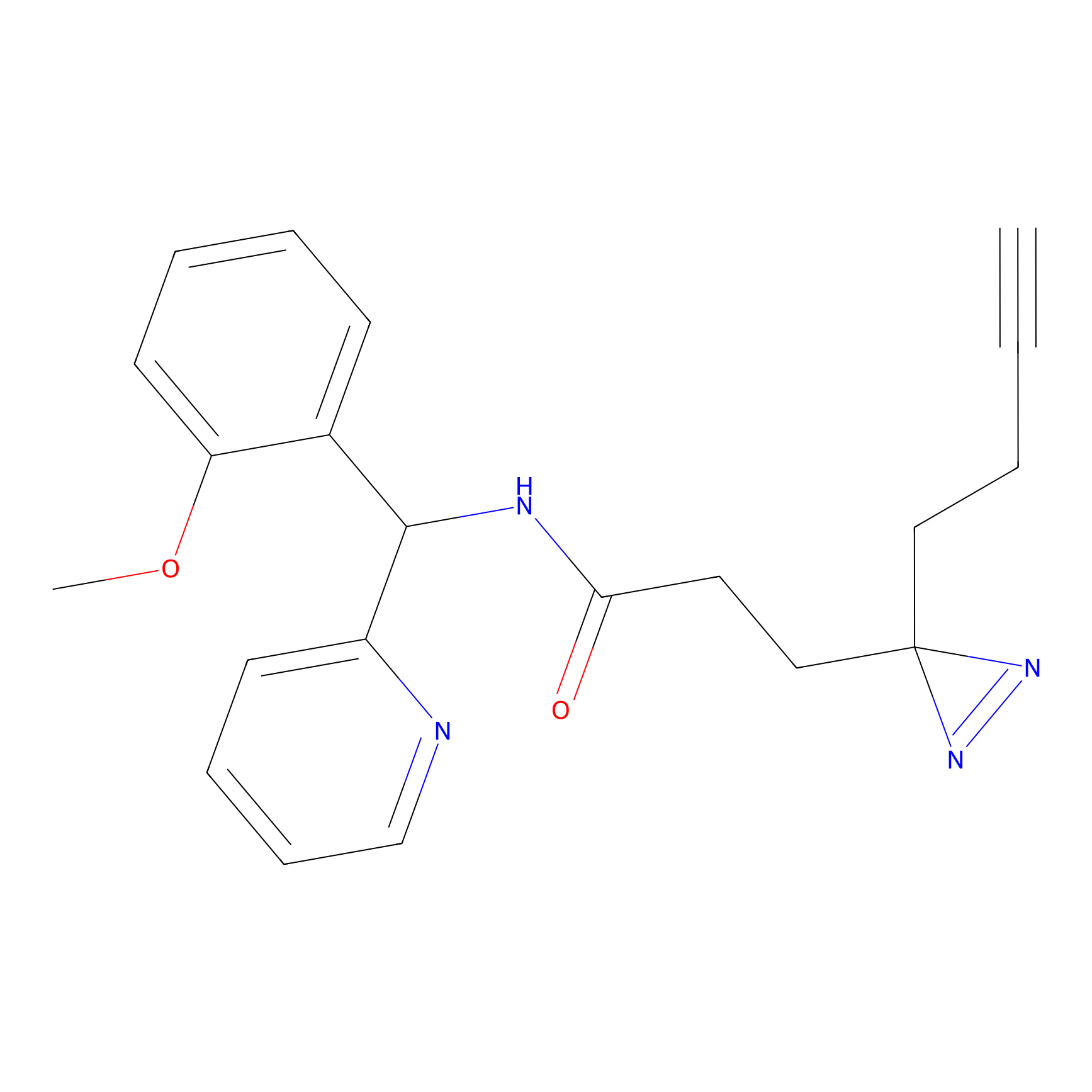 |
5.66 | LDD1869 | [19] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C62(0.40) | LDD2142 | [3] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C62(0.95) | LDD2117 | [3] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C62(1.12) | LDD2152 | [3] |
| LDCM0214 | AC1 | HCT 116 | C62(0.76) | LDD0531 | [6] |
| LDCM0279 | AC2 | HCT 116 | C62(0.94) | LDD0596 | [6] |
| LDCM0290 | AC3 | HCT 116 | C62(0.96) | LDD0607 | [6] |
| LDCM0301 | AC4 | HCT 116 | C62(0.97) | LDD0618 | [6] |
| LDCM0312 | AC5 | HCT 116 | C62(0.98) | LDD0629 | [6] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [17] |
| LDCM0367 | CL1 | HCT 116 | C62(0.99) | LDD0684 | [6] |
| LDCM0368 | CL10 | HCT 116 | C62(0.69) | LDD0685 | [6] |
| LDCM0369 | CL100 | HCT 116 | C62(0.91) | LDD0686 | [6] |
| LDCM0379 | CL11 | HCT 116 | C62(0.85) | LDD0696 | [6] |
| LDCM0390 | CL12 | HCT 116 | C62(1.06) | LDD0707 | [6] |
| LDCM0400 | CL13 | HCT 116 | C62(0.78) | LDD0717 | [6] |
| LDCM0401 | CL14 | HCT 116 | C62(0.87) | LDD0718 | [6] |
| LDCM0402 | CL15 | HCT 116 | C62(0.52) | LDD0719 | [6] |
| LDCM0407 | CL2 | HCT 116 | C62(0.93) | LDD0724 | [6] |
| LDCM0418 | CL3 | HCT 116 | C62(0.88) | LDD0735 | [6] |
| LDCM0420 | CL31 | HCT 116 | C62(1.12) | LDD0737 | [6] |
| LDCM0421 | CL32 | HCT 116 | C62(1.15) | LDD0738 | [6] |
| LDCM0422 | CL33 | HCT 116 | C62(0.97) | LDD0739 | [6] |
| LDCM0423 | CL34 | HCT 116 | C62(1.07) | LDD0740 | [6] |
| LDCM0424 | CL35 | HCT 116 | C62(1.00) | LDD0741 | [6] |
| LDCM0425 | CL36 | HCT 116 | C62(0.92) | LDD0742 | [6] |
| LDCM0426 | CL37 | HCT 116 | C62(1.26) | LDD0743 | [6] |
| LDCM0428 | CL39 | HCT 116 | C62(1.18) | LDD0745 | [6] |
| LDCM0429 | CL4 | HCT 116 | C62(0.85) | LDD0746 | [6] |
| LDCM0430 | CL40 | HCT 116 | C62(1.05) | LDD0747 | [6] |
| LDCM0431 | CL41 | HCT 116 | C62(1.20) | LDD0748 | [6] |
| LDCM0432 | CL42 | HCT 116 | C62(0.96) | LDD0749 | [6] |
| LDCM0433 | CL43 | HCT 116 | C62(1.06) | LDD0750 | [6] |
| LDCM0434 | CL44 | HCT 116 | C62(1.20) | LDD0751 | [6] |
| LDCM0435 | CL45 | HCT 116 | C62(0.98) | LDD0752 | [6] |
| LDCM0440 | CL5 | HCT 116 | C62(0.93) | LDD0757 | [6] |
| LDCM0451 | CL6 | HCT 116 | C62(0.70) | LDD0768 | [6] |
| LDCM0453 | CL61 | HCT 116 | C62(0.94) | LDD0770 | [6] |
| LDCM0454 | CL62 | HCT 116 | C62(0.96) | LDD0771 | [6] |
| LDCM0455 | CL63 | HCT 116 | C62(0.88) | LDD0772 | [6] |
| LDCM0456 | CL64 | HCT 116 | C62(0.89) | LDD0773 | [6] |
| LDCM0457 | CL65 | HCT 116 | C62(0.96) | LDD0774 | [6] |
| LDCM0458 | CL66 | HCT 116 | C62(1.00) | LDD0775 | [6] |
| LDCM0459 | CL67 | HCT 116 | C62(0.91) | LDD0776 | [6] |
| LDCM0460 | CL68 | HCT 116 | C62(0.75) | LDD0777 | [6] |
| LDCM0461 | CL69 | HCT 116 | C62(0.75) | LDD0778 | [6] |
| LDCM0462 | CL7 | HCT 116 | C62(0.87) | LDD0779 | [6] |
| LDCM0463 | CL70 | HCT 116 | C62(0.74) | LDD0780 | [6] |
| LDCM0464 | CL71 | HCT 116 | C62(0.85) | LDD0781 | [6] |
| LDCM0465 | CL72 | HCT 116 | C62(1.06) | LDD0782 | [6] |
| LDCM0466 | CL73 | HCT 116 | C62(0.88) | LDD0783 | [6] |
| LDCM0467 | CL74 | HCT 116 | C62(0.89) | LDD0784 | [6] |
| LDCM0473 | CL8 | HCT 116 | C62(0.94) | LDD0790 | [6] |
| LDCM0484 | CL9 | HCT 116 | C62(0.89) | LDD0801 | [6] |
| LDCM0486 | CL91 | HCT 116 | C62(0.95) | LDD0803 | [6] |
| LDCM0487 | CL92 | HCT 116 | C62(0.87) | LDD0804 | [6] |
| LDCM0488 | CL93 | HCT 116 | C62(0.92) | LDD0805 | [6] |
| LDCM0489 | CL94 | HCT 116 | C62(0.92) | LDD0806 | [6] |
| LDCM0490 | CL95 | HCT 116 | C62(1.04) | LDD0807 | [6] |
| LDCM0491 | CL96 | HCT 116 | C62(0.89) | LDD0808 | [6] |
| LDCM0492 | CL97 | HCT 116 | C62(0.91) | LDD0809 | [6] |
| LDCM0493 | CL98 | HCT 116 | C62(0.97) | LDD0810 | [6] |
| LDCM0494 | CL99 | HCT 116 | C62(0.93) | LDD0811 | [6] |
| LDCM0468 | Fragment33 | HCT 116 | C62(0.93) | LDD0785 | [6] |
| LDCM0427 | Fragment51 | HCT 116 | C62(0.51) | LDD0744 | [6] |
| LDCM0116 | HHS-0101 | DM93 | Y87(0.96); Y38(1.20); Y16(1.27) | LDD0264 | [4] |
| LDCM0117 | HHS-0201 | DM93 | Y38(0.74); Y87(0.92); Y16(1.01) | LDD0265 | [4] |
| LDCM0118 | HHS-0301 | DM93 | Y38(0.40); Y87(0.86); Y16(0.93) | LDD0266 | [4] |
| LDCM0119 | HHS-0401 | DM93 | Y38(0.49); Y87(1.13); Y16(1.30) | LDD0267 | [4] |
| LDCM0120 | HHS-0701 | DM93 | Y38(0.71); Y16(0.75); Y87(0.83) | LDD0268 | [4] |
| LDCM0022 | KB02 | HCC1954 | C62(1.30) | LDD2343 | [20] |
| LDCM0023 | KB03 | MDA-MB-231 | C62(0.80) | LDD1701 | [3] |
| LDCM0024 | KB05 | HCC1954 | C62(1.48) | LDD3177 | [20] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C62(1.06) | LDD2093 | [3] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C62(0.82) | LDD2098 | [3] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C62(1.13) | LDD2099 | [3] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C62(0.88) | LDD2101 | [3] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C62(0.39) | LDD2104 | [3] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C62(0.52) | LDD2106 | [3] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C62(1.23) | LDD2107 | [3] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C62(0.60) | LDD2108 | [3] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C62(1.29) | LDD2109 | [3] |
| LDCM0517 | Nucleophilic fragment 21b | MDA-MB-231 | C62(0.42) | LDD2110 | [3] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C62(1.10) | LDD2111 | [3] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C62(0.45) | LDD2115 | [3] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C62(2.10) | LDD2119 | [3] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C62(0.87) | LDD2125 | [3] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C62(1.05) | LDD2127 | [3] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C62(1.07) | LDD2129 | [3] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C62(0.50) | LDD2134 | [3] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C62(1.37) | LDD2135 | [3] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C62(0.98) | LDD2137 | [3] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C62(1.95) | LDD1700 | [3] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C62(0.76) | LDD2140 | [3] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C62(2.70) | LDD2144 | [3] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C62(0.96) | LDD2146 | [3] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Telomeric repeat-binding factor 2-interacting protein 1 (TERF2IP) | RAP1 family | Q9NYB0 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Testin (TES) | Prickle / espinas / testin family | Q9UGI8 | |||
| Protocadherin Fat 1 (FAT1) | . | Q14517 | |||
References
