Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
8.32 | LDD0402 | [1] | |
|
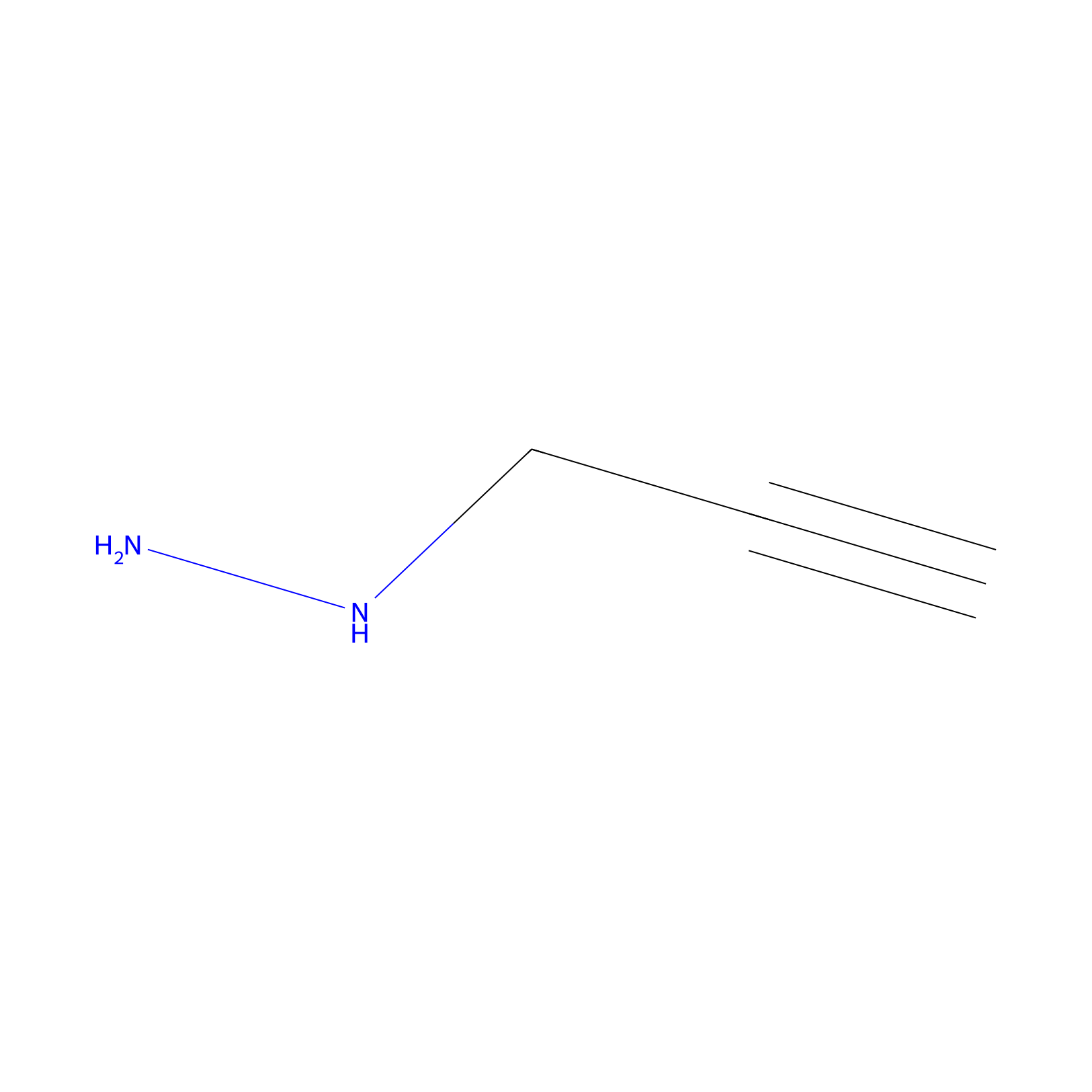
Alkylaryl probe 2 Probe Info |
 |
10.00 | LDD0391 | [2] | |
|
STPyne Probe Info |
 |
K383(7.26); K386(6.46); K404(6.71); K434(5.55) | LDD0277 | [3] | |
|
OPA-S-S-alkyne Probe Info |
 |
K337(1.70) | LDD3494 | [4] | |
|
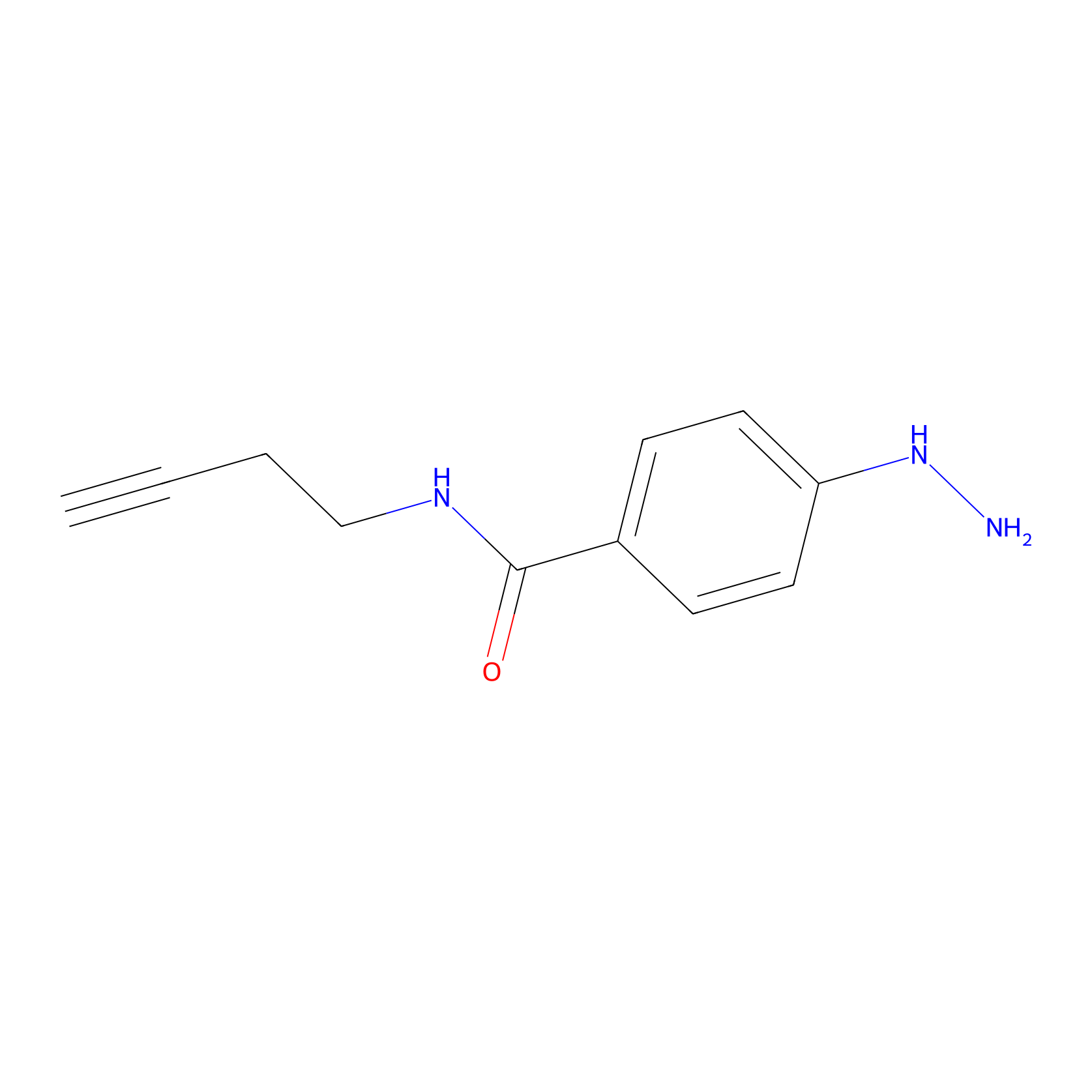
Alkylaryl probe 1 Probe Info |
 |
13.00 | LDD0388 | [2] | |
|
DBIA Probe Info |
 |
C574(5.17) | LDD3310 | [5] | |
|
JZ128-DTB Probe Info |
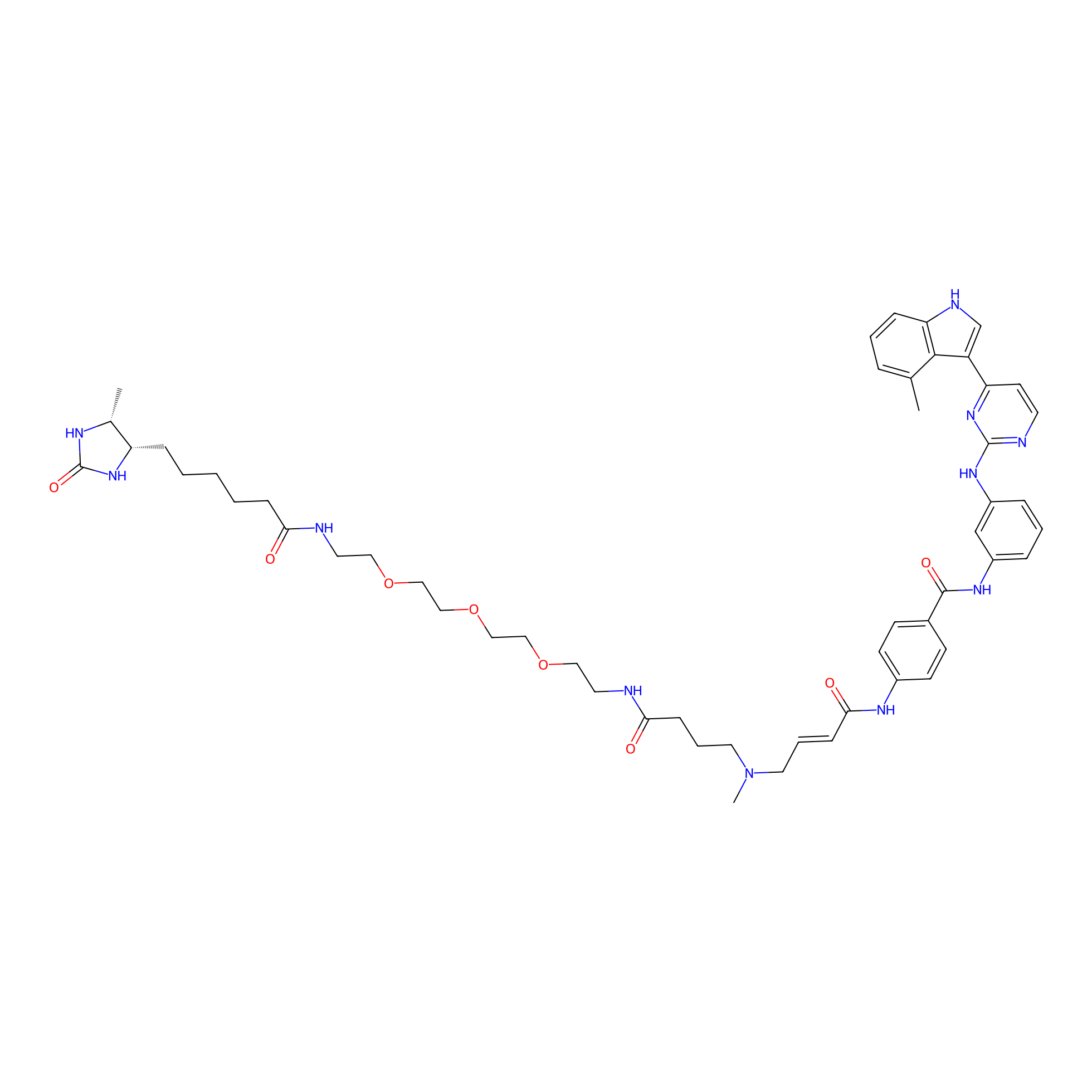 |
N.A. | LDD0462 | [6] | |
|
BTD Probe Info |
 |
C574(1.47) | LDD1700 | [7] | |
|
AHL-Pu-1 Probe Info |
 |
C574(2.71) | LDD0171 | [8] | |
|
IPM Probe Info |
 |
C574(49.39) | LDD1701 | [7] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [9] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C574(0.97) | LDD2112 | [7] |
| LDCM0026 | 4SU-RNA+native RNA | DM93 | C574(2.71) | LDD0171 | [8] |
| LDCM0156 | Aniline | NCI-H1299 | 7.12 | LDD0403 | [1] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C574(50.23) | LDD1702 | [7] |
| LDCM0179 | JZ128 | PC-3 | N.A. | LDD0462 | [6] |
| LDCM0022 | KB02 | 42-MG-BA | C127(1.90) | LDD2244 | [5] |
| LDCM0023 | KB03 | MDA-MB-231 | C574(49.39) | LDD1701 | [7] |
| LDCM0024 | KB05 | COLO792 | C574(5.17) | LDD3310 | [5] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C574(1.49) | LDD2092 | [7] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C574(1.02) | LDD2093 | [7] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C574(2.21) | LDD2094 | [7] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C574(1.00) | LDD2097 | [7] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C574(1.04) | LDD2098 | [7] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C574(1.92); C93(0.77) | LDD2100 | [7] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C574(1.53); C93(1.71) | LDD2104 | [7] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C574(0.73) | LDD2108 | [7] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C574(0.79) | LDD2109 | [7] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C574(1.11) | LDD2111 | [7] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C574(2.17) | LDD2119 | [7] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C574(0.92) | LDD2120 | [7] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C574(1.09) | LDD2127 | [7] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C574(1.00) | LDD2128 | [7] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C574(0.87) | LDD2129 | [7] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C574(0.67) | LDD2133 | [7] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C574(0.48) | LDD2134 | [7] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C574(0.89) | LDD2135 | [7] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C574(1.85) | LDD2136 | [7] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C574(1.47) | LDD1700 | [7] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C574(0.63) | LDD2140 | [7] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C574(0.93) | LDD2143 | [7] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C574(0.99) | LDD2146 | [7] |
| LDCM0553 | Nucleophilic fragment 6b | MDA-MB-231 | C574(1.96) | LDD2147 | [7] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C574(0.59) | LDD2148 | [7] |
| LDCM0099 | Phenelzine | MDA-MB-231 | 13.00 | LDD0388 | [2] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Stomatin (STOM) | Band 7/mec-2 family | P27105 | |||
| Aquaporin-6 (AQP6) | MIP/aquaporin (TC 1.A.8) family | Q13520 | |||
Immunoglobulin
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Junctional adhesion molecule A (F11R) | Immunoglobulin superfamily | Q9Y624 | |||
Other
The Drug(s) Related To This Target
Phase 3
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Epigallocatechin Gallate | Small molecular drug | D0U2BH | |||
Phase 2
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Bt1718 | Bicycle toxin conjugate | DE2Z7V | |||
Investigative
Discontinued
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Gm6001 | Small molecular drug | D0T9HG | |||
References
