Details of the Target
General Information of Target
| Target ID | LDTP03354 | |||||
|---|---|---|---|---|---|---|
| Target Name | Large ribosomal subunit protein eL13 (RPL13) | |||||
| Gene Name | RPL13 | |||||
| Gene ID | 6137 | |||||
| Synonyms |
BBC1; Large ribosomal subunit protein eL13; 60S ribosomal protein L13; Breast basic conserved protein 1 |
|||||
| 3D Structure | ||||||
| Sequence |
MAPSRNGMVLKPHFHKDWQRRVATWFNQPARKIRRRKARQAKARRIAPRPASGPIRPIVR
CPTVRYHTKVRAGRGFSLEELRVAGIHKKVARTIGISVDPRRRNKSTESLQANVQRLKEY RSKLILFPRKPSAPKKGDSSAEELKLATQLTGPVMPVRNVYKKEKARVITEEEKNFKAFA SLRMARANARLFGIRAKRAKEAAEQDVEKKK |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Eukaryotic ribosomal protein eL13 family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Component of the ribosome, a large ribonucleoprotein complex responsible for the synthesis of proteins in the cell. The small ribosomal subunit (SSU) binds messenger RNAs (mRNAs) and translates the encoded message by selecting cognate aminoacyl-transfer RNA (tRNA) molecules (Probable). The large subunit (LSU) contains the ribosomal catalytic site termed the peptidyl transferase center (PTC), which catalyzes the formation of peptide bonds, thereby polymerizing the amino acids delivered by tRNAs into a polypeptide chain (Probable). The nascent polypeptides leave the ribosome through a tunnel in the LSU and interact with protein factors that function in enzymatic processing, targeting, and the membrane insertion of nascent chains at the exit of the ribosomal tunnel (Probable). As part of the LSU, it is probably required for its formation and the maturation of rRNAs. Plays a role in bone development.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| IM95 | SNV: p.R186C | DBIA Probe Info | |||
| JHH1 | SNV: p.N113S | . | |||
| NCIH2172 | SNV: p.F179L | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
8.44 | LDD0402 | [1] | |
|
A-EBA Probe Info |
 |
3.15 | LDD0215 | [2] | |
|
C-Sul Probe Info |
 |
4.78 | LDD0066 | [3] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [4] | |
|
STPyne Probe Info |
 |
K123(8.50); K145(7.63); K163(1.10); K174(6.25) | LDD0277 | [5] | |
|
ONAyne Probe Info |
 |
K177(0.00); K69(0.00); K200(0.00); K105(0.00) | LDD0273 | [5] | |
|
AF-1 Probe Info |
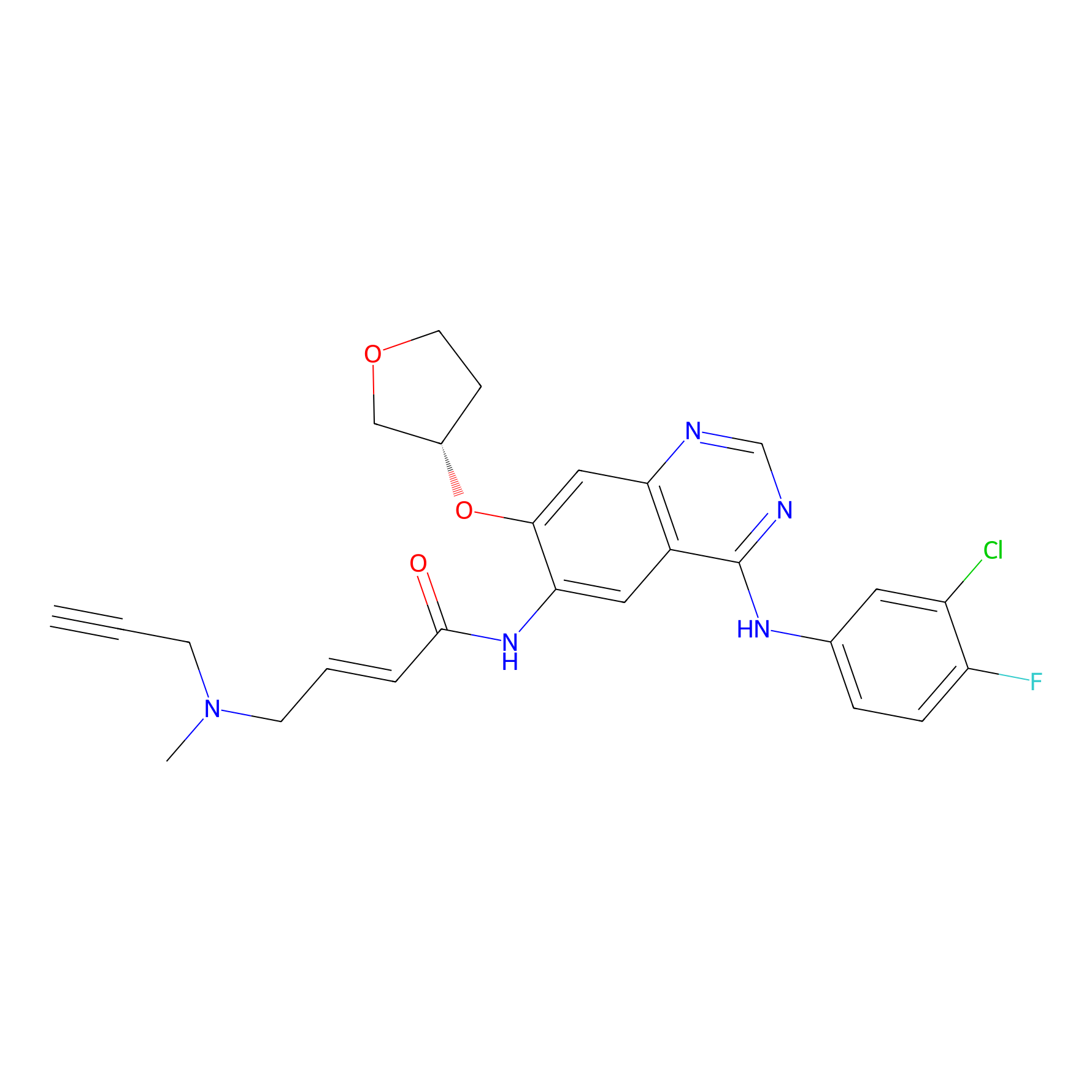 |
1.54 | LDD0421 | [6] | |
|
DBIA Probe Info |
 |
C186(1.85) | LDD2356 | [7] | |
|
AZ-9 Probe Info |
 |
8.02 | LDD2154 | [8] | |
|
W1 Probe Info |
 |
S122(0.94) | LDD0239 | [9] | |
|
AMP probe Probe Info |
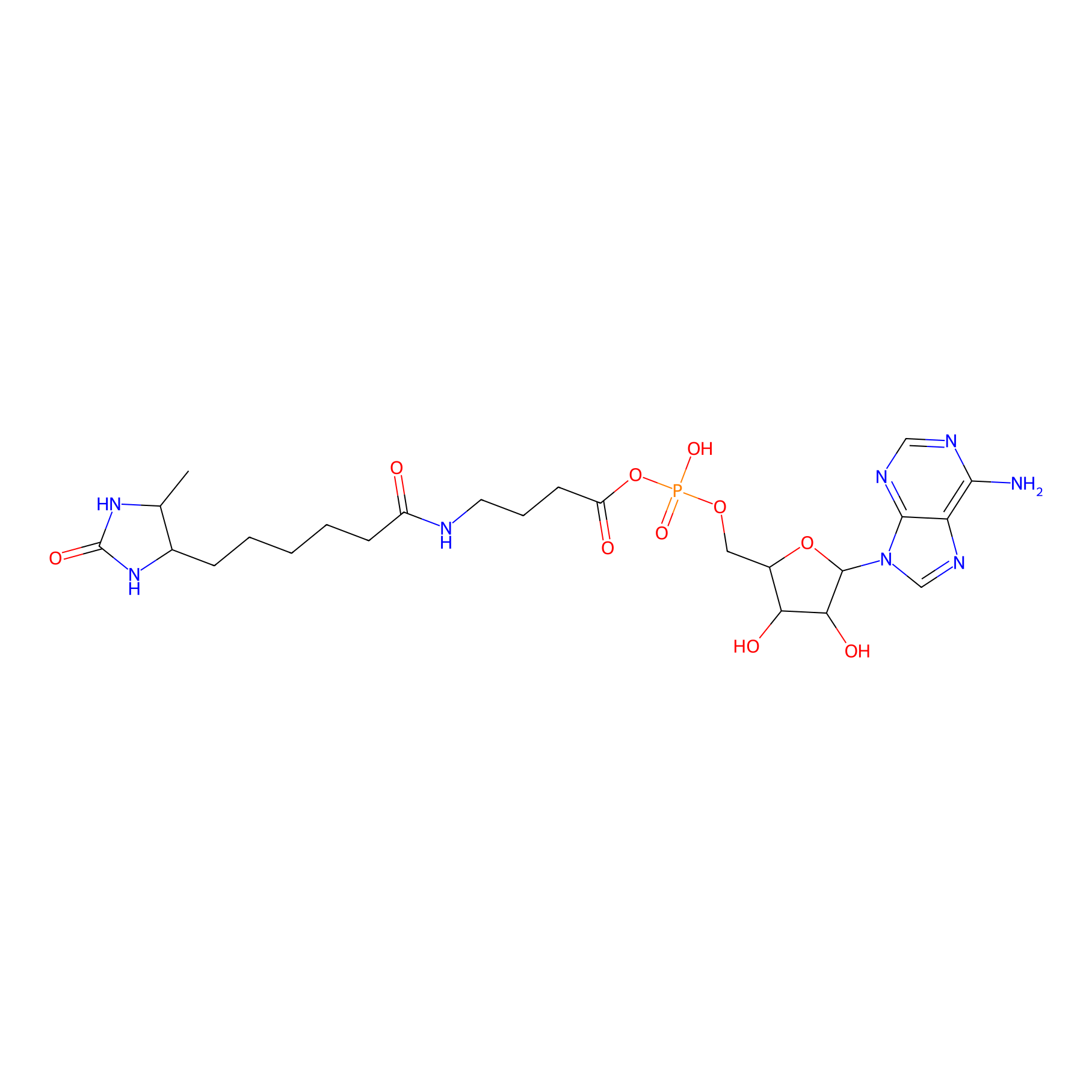 |
N.A. | LDD0200 | [10] | |
|
ATP probe Probe Info |
 |
K200(0.00); K136(0.00); K174(0.00); K177(0.00) | LDD0199 | [10] | |
|
ATP probe Probe Info |
 |
K136(0.00); K200(0.00); K11(0.00) | LDD0035 | [11] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0356 | [12] | |
|
NHS Probe Info |
 |
K177(0.00); K145(0.00) | LDD0010 | [13] | |
|
OSF Probe Info |
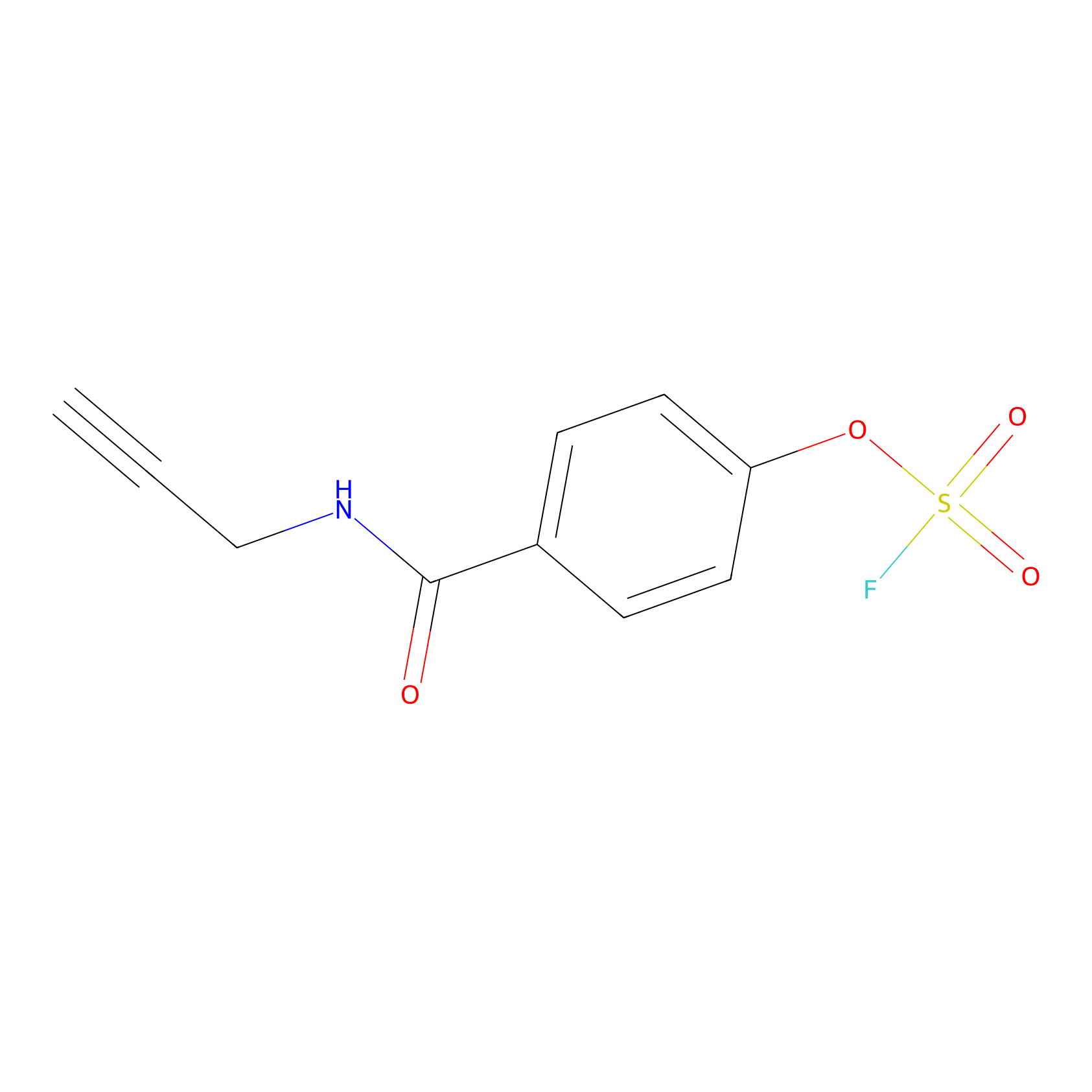 |
N.A. | LDD0029 | [14] | |
|
SF Probe Info |
 |
K88(0.00); K130(0.00); K123(0.00); K177(0.00) | LDD0028 | [14] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [15] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [16] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [12] | |
|
Acrolein Probe Info |
 |
H13(0.00); H15(0.00) | LDD0217 | [17] | |
|
AOyne Probe Info |
 |
6.10 | LDD0443 | [18] | |
|
MPP-AC Probe Info |
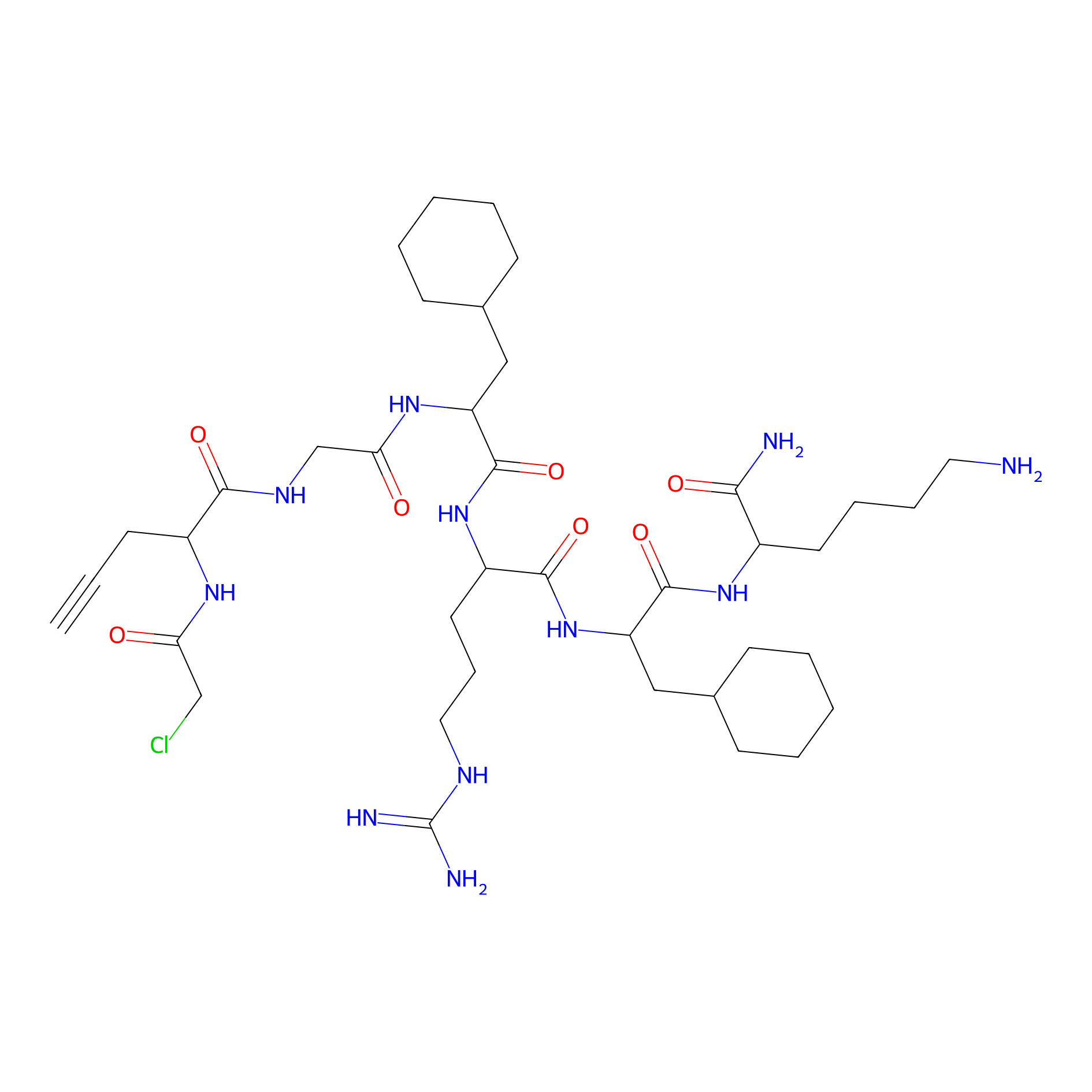 |
N.A. | LDD0428 | [19] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DA-2 Probe Info |
 |
N.A. | LDD0072 | [20] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0166 | Afatinib | A431 | 1.54 | LDD0421 | [6] |
| LDCM0151 | AZ-11 | HeLa | 8.02 | LDD2154 | [8] |
| LDCM0108 | Chloroacetamide | HeLa | H13(0.00); H15(0.00) | LDD0222 | [17] |
| LDCM0107 | IAA | HeLa | H13(0.00); H15(0.00) | LDD0221 | [17] |
| LDCM0022 | KB02 | HGC-27 | C186(1.85) | LDD2356 | [7] |
| LDCM0023 | KB03 | HGC-27 | C186(1.82) | LDD2773 | [7] |
| LDCM0024 | KB05 | HGC-27 | C186(2.06) | LDD3190 | [7] |
| LDCM0109 | NEM | HeLa | H13(0.00); H15(0.00) | LDD0223 | [17] |
| LDCM0112 | W16 | Hep-G2 | S122(0.94) | LDD0239 | [9] |
| LDCM0113 | W17 | Hep-G2 | S122(0.59); K123(0.62) | LDD0240 | [9] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Leucine-rich repeat serine/threonine-protein kinase 2 (LRRK2) | TKL Ser/Thr protein kinase family | Q5S007 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Huntingtin (HTT) | Huntingtin family | P42858 | |||
Other
References
