Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
IPM Probe Info |
 |
C280(2.34) | LDD1702 | [1] | |
|
DBIA Probe Info |
 |
C185(1.17) | LDD0532 | [2] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0166 | [3] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [4] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [5] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
IMP2070 Probe Info |
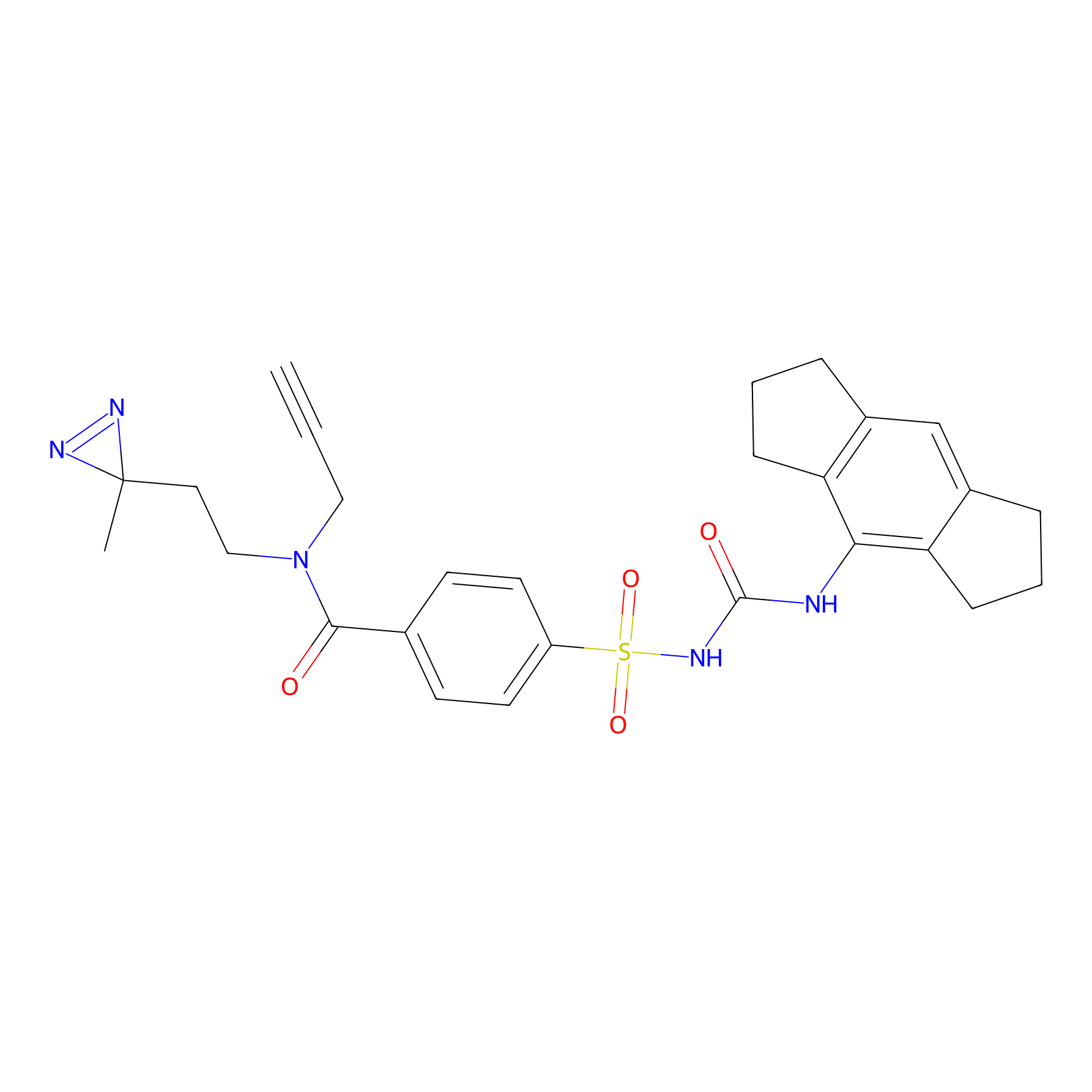 |
7.29 | LDD0280 | [6] | |
|
C187 Probe Info |
 |
58.08 | LDD1865 | [7] | |
|
C191 Probe Info |
 |
32.45 | LDD1868 | [7] | |
|
C193 Probe Info |
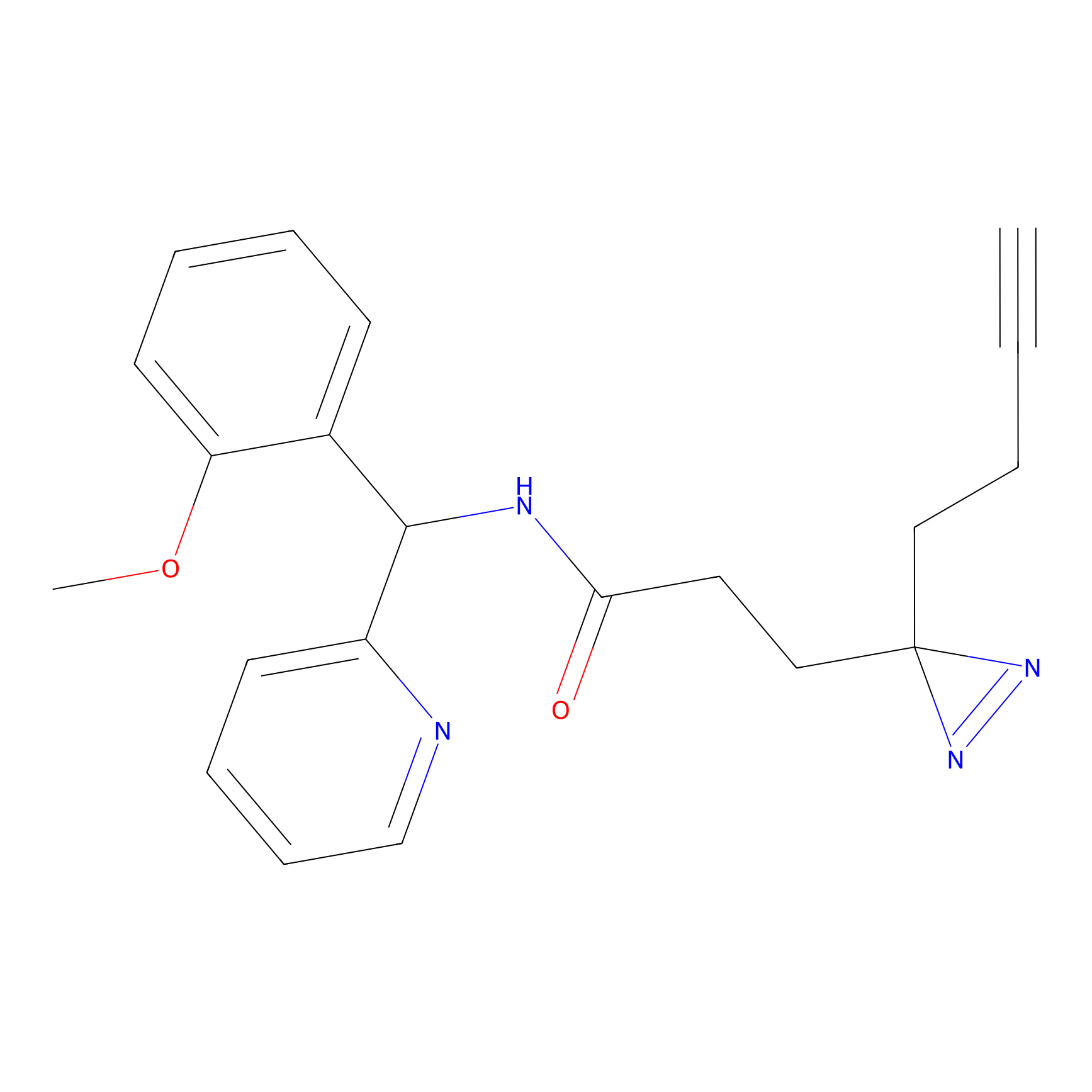 |
12.47 | LDD1869 | [7] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0215 | AC10 | HCT 116 | C185(1.17) | LDD0532 | [2] |
| LDCM0226 | AC11 | HCT 116 | C185(0.98) | LDD0543 | [2] |
| LDCM0237 | AC12 | HCT 116 | C185(0.90) | LDD0554 | [2] |
| LDCM0246 | AC128 | HCT 116 | C185(1.21) | LDD0563 | [2] |
| LDCM0247 | AC129 | HCT 116 | C185(1.25) | LDD0564 | [2] |
| LDCM0249 | AC130 | HCT 116 | C185(1.11) | LDD0566 | [2] |
| LDCM0250 | AC131 | HCT 116 | C185(1.02) | LDD0567 | [2] |
| LDCM0251 | AC132 | HCT 116 | C185(0.89) | LDD0568 | [2] |
| LDCM0252 | AC133 | HCT 116 | C185(1.10) | LDD0569 | [2] |
| LDCM0253 | AC134 | HCT 116 | C185(1.17) | LDD0570 | [2] |
| LDCM0254 | AC135 | HCT 116 | C185(1.19) | LDD0571 | [2] |
| LDCM0255 | AC136 | HCT 116 | C185(0.96) | LDD0572 | [2] |
| LDCM0256 | AC137 | HCT 116 | C185(1.00) | LDD0573 | [2] |
| LDCM0257 | AC138 | HCT 116 | C185(0.95) | LDD0574 | [2] |
| LDCM0258 | AC139 | HCT 116 | C185(1.39) | LDD0575 | [2] |
| LDCM0259 | AC14 | HCT 116 | C185(1.09) | LDD0576 | [2] |
| LDCM0260 | AC140 | HCT 116 | C185(1.33) | LDD0577 | [2] |
| LDCM0261 | AC141 | HCT 116 | C185(1.10) | LDD0578 | [2] |
| LDCM0262 | AC142 | HCT 116 | C185(1.13) | LDD0579 | [2] |
| LDCM0270 | AC15 | HCT 116 | C185(0.94) | LDD0587 | [2] |
| LDCM0276 | AC17 | HCT 116 | C185(1.20) | LDD0593 | [2] |
| LDCM0277 | AC18 | HCT 116 | C185(1.29) | LDD0594 | [2] |
| LDCM0278 | AC19 | HCT 116 | C185(1.26) | LDD0595 | [2] |
| LDCM0280 | AC20 | HCT 116 | C185(1.19) | LDD0597 | [2] |
| LDCM0281 | AC21 | HCT 116 | C185(1.21) | LDD0598 | [2] |
| LDCM0282 | AC22 | HCT 116 | C185(1.12) | LDD0599 | [2] |
| LDCM0283 | AC23 | HCT 116 | C185(1.32) | LDD0600 | [2] |
| LDCM0284 | AC24 | HCT 116 | C185(1.18) | LDD0601 | [2] |
| LDCM0323 | AC6 | HCT 116 | C185(1.02) | LDD0640 | [2] |
| LDCM0334 | AC7 | HCT 116 | C185(1.02) | LDD0651 | [2] |
| LDCM0345 | AC8 | HCT 116 | C185(1.11) | LDD0662 | [2] |
| LDCM0248 | AKOS034007472 | HCT 116 | C185(1.10) | LDD0565 | [2] |
| LDCM0356 | AKOS034007680 | HCT 116 | C185(1.07) | LDD0673 | [2] |
| LDCM0275 | AKOS034007705 | HCT 116 | C185(1.34) | LDD0592 | [2] |
| LDCM0370 | CL101 | HCT 116 | C185(1.03) | LDD0687 | [2] |
| LDCM0371 | CL102 | HCT 116 | C185(0.95) | LDD0688 | [2] |
| LDCM0372 | CL103 | HCT 116 | C185(0.92) | LDD0689 | [2] |
| LDCM0373 | CL104 | HCT 116 | C185(0.96) | LDD0690 | [2] |
| LDCM0374 | CL105 | HCT 116 | C185(1.29) | LDD0691 | [2] |
| LDCM0375 | CL106 | HCT 116 | C185(1.47) | LDD0692 | [2] |
| LDCM0376 | CL107 | HCT 116 | C185(1.37) | LDD0693 | [2] |
| LDCM0377 | CL108 | HCT 116 | C185(1.56) | LDD0694 | [2] |
| LDCM0378 | CL109 | HCT 116 | C185(1.37) | LDD0695 | [2] |
| LDCM0380 | CL110 | HCT 116 | C185(1.49) | LDD0697 | [2] |
| LDCM0381 | CL111 | HCT 116 | C185(1.20) | LDD0698 | [2] |
| LDCM0436 | CL46 | HCT 116 | C185(0.77) | LDD0753 | [2] |
| LDCM0437 | CL47 | HCT 116 | C185(1.04) | LDD0754 | [2] |
| LDCM0438 | CL48 | HCT 116 | C185(0.72) | LDD0755 | [2] |
| LDCM0439 | CL49 | HCT 116 | C185(0.77) | LDD0756 | [2] |
| LDCM0441 | CL50 | HCT 116 | C185(0.93) | LDD0758 | [2] |
| LDCM0442 | CL51 | HCT 116 | C185(0.78) | LDD0759 | [2] |
| LDCM0443 | CL52 | HCT 116 | C185(0.73) | LDD0760 | [2] |
| LDCM0444 | CL53 | HCT 116 | C185(0.72) | LDD0761 | [2] |
| LDCM0445 | CL54 | HCT 116 | C185(0.70) | LDD0762 | [2] |
| LDCM0446 | CL55 | HCT 116 | C185(0.71) | LDD0763 | [2] |
| LDCM0447 | CL56 | HCT 116 | C185(0.76) | LDD0764 | [2] |
| LDCM0448 | CL57 | HCT 116 | C185(0.72) | LDD0765 | [2] |
| LDCM0449 | CL58 | HCT 116 | C185(0.70) | LDD0766 | [2] |
| LDCM0450 | CL59 | HCT 116 | C185(0.90) | LDD0767 | [2] |
| LDCM0452 | CL60 | HCT 116 | C185(0.96) | LDD0769 | [2] |
| LDCM0453 | CL61 | HCT 116 | C185(0.93) | LDD0770 | [2] |
| LDCM0454 | CL62 | HCT 116 | C185(1.08) | LDD0771 | [2] |
| LDCM0455 | CL63 | HCT 116 | C185(1.05) | LDD0772 | [2] |
| LDCM0456 | CL64 | HCT 116 | C185(1.14) | LDD0773 | [2] |
| LDCM0457 | CL65 | HCT 116 | C185(1.00) | LDD0774 | [2] |
| LDCM0458 | CL66 | HCT 116 | C185(1.10) | LDD0775 | [2] |
| LDCM0459 | CL67 | HCT 116 | C185(1.16) | LDD0776 | [2] |
| LDCM0460 | CL68 | HCT 116 | C185(1.00) | LDD0777 | [2] |
| LDCM0461 | CL69 | HCT 116 | C185(1.31) | LDD0778 | [2] |
| LDCM0463 | CL70 | HCT 116 | C185(1.03) | LDD0780 | [2] |
| LDCM0464 | CL71 | HCT 116 | C185(1.07) | LDD0781 | [2] |
| LDCM0465 | CL72 | HCT 116 | C185(1.09) | LDD0782 | [2] |
| LDCM0466 | CL73 | HCT 116 | C185(1.13) | LDD0783 | [2] |
| LDCM0467 | CL74 | HCT 116 | C185(0.99) | LDD0784 | [2] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C280(2.34) | LDD1702 | [1] |
| LDCM0468 | Fragment33 | HCT 116 | C185(1.27) | LDD0785 | [2] |
| LDCM0022 | KB02 | CMK | C185(2.02) | LDD2302 | [8] |
| LDCM0023 | KB03 | EoL-1 | C280(2.53) | LDD2741 | [8] |
| LDCM0024 | KB05 | SKMEL24 | C280(2.03) | LDD3323 | [8] |
The Interaction Atlas With This Target
References
