Details of the Target
General Information of Target
| Target ID | LDTP01165 | |||||
|---|---|---|---|---|---|---|
| Target Name | NADH dehydrogenase iron-sulfur protein 7, mitochondrial (NDUFS7) | |||||
| Gene Name | NDUFS7 | |||||
| Gene ID | 374291 | |||||
| Synonyms |
NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial; EC 7.1.1.2; Complex I-20kD; CI-20kD; NADH-ubiquinone oxidoreductase 20 kDa subunit; PSST subunit |
|||||
| 3D Structure | ||||||
| Sequence |
MAVLSAPGLRGFRILGLRSSVGPAVQARGVHQSVATDGPSSTQPALPKARAVAPKPSSRG
EYVVAKLDDLVNWARRSSLWPMTFGLACCAVEMMHMAAPRYDMDRFGVVFRASPRQSDVM IVAGTLTNKMAPALRKVYDQMPEPRYVVSMGSCANGGGYYHYSYSVVRGCDRIVPVDIYI PGCPPTAEALLYGILQLQRKIKRERRLQIWYRR |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Complex I 20 kDa subunit family
|
|||||
| Subcellular location |
Mitochondrion inner membrane
|
|||||
| Function |
Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) which catalyzes electron transfer from NADH through the respiratory chain, using ubiquinone as an electron acceptor. Essential for the catalytic activity of complex I.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TH211 Probe Info |
 |
Y62(16.45) | LDD0257 | [1] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [2] | |
|
Probe 1 Probe Info |
 |
Y62(23.80) | LDD3495 | [3] | |
|
DBIA Probe Info |
 |
C183(1.25) | LDD3342 | [4] | |
|
Jackson_14 Probe Info |
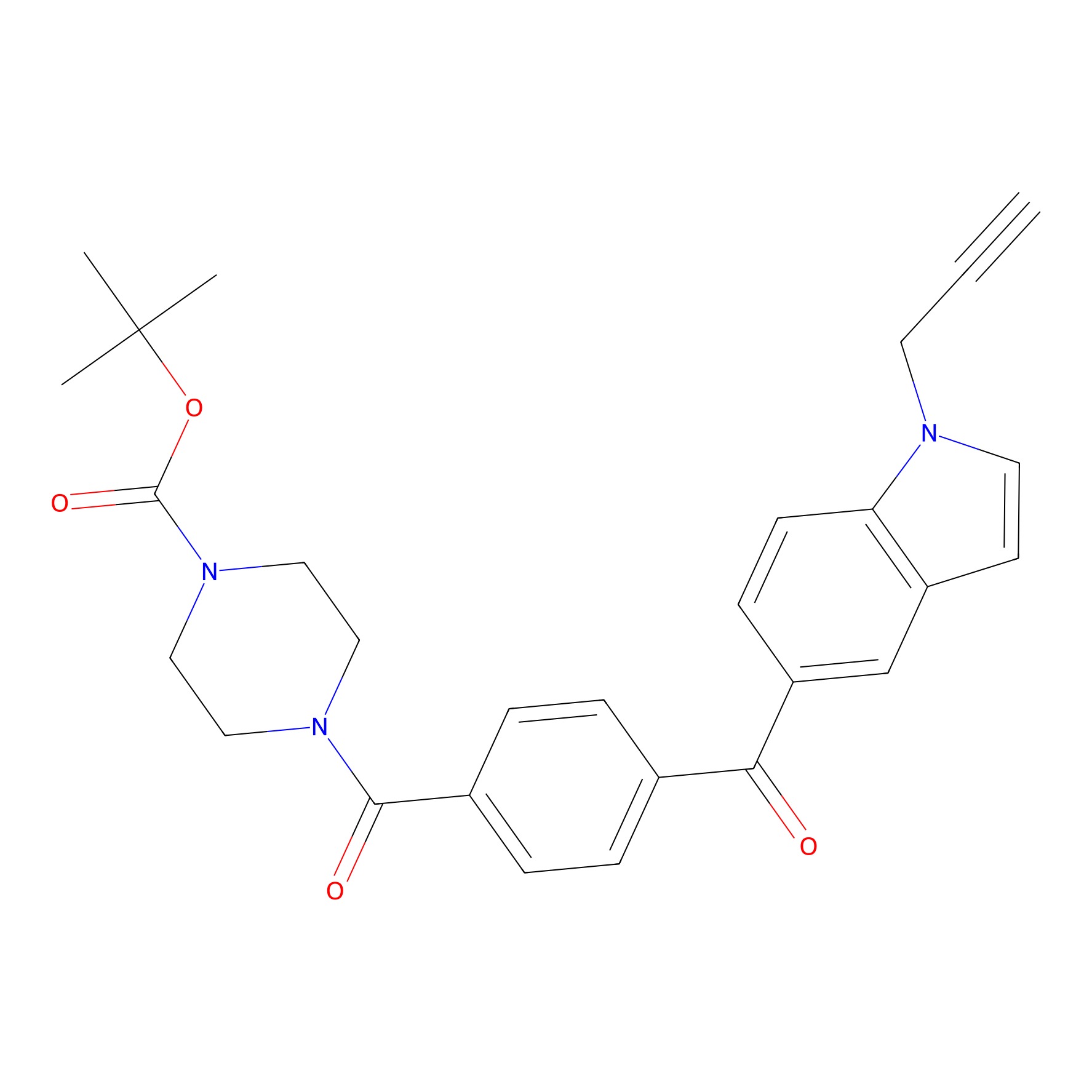 |
5.88 | LDD0123 | [5] | |
|
IPM Probe Info |
 |
C194(4.56) | LDD0406 | [6] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0358 | [7] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [8] | |
|
IA-alkyne Probe Info |
 |
C183(0.00); C88(0.00); C89(0.00) | LDD0036 | [8] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [8] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [9] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [10] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [10] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [11] | |
|
1c-yne Probe Info |
 |
K136(0.00); K66(0.00) | LDD0228 | [7] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [12] | |
|
AOyne Probe Info |
 |
11.10 | LDD0443 | [13] | |
|
NAIA_5 Probe Info |
 |
C183(0.00); C153(0.00) | LDD2223 | [9] | |
|
TER-AC Probe Info |
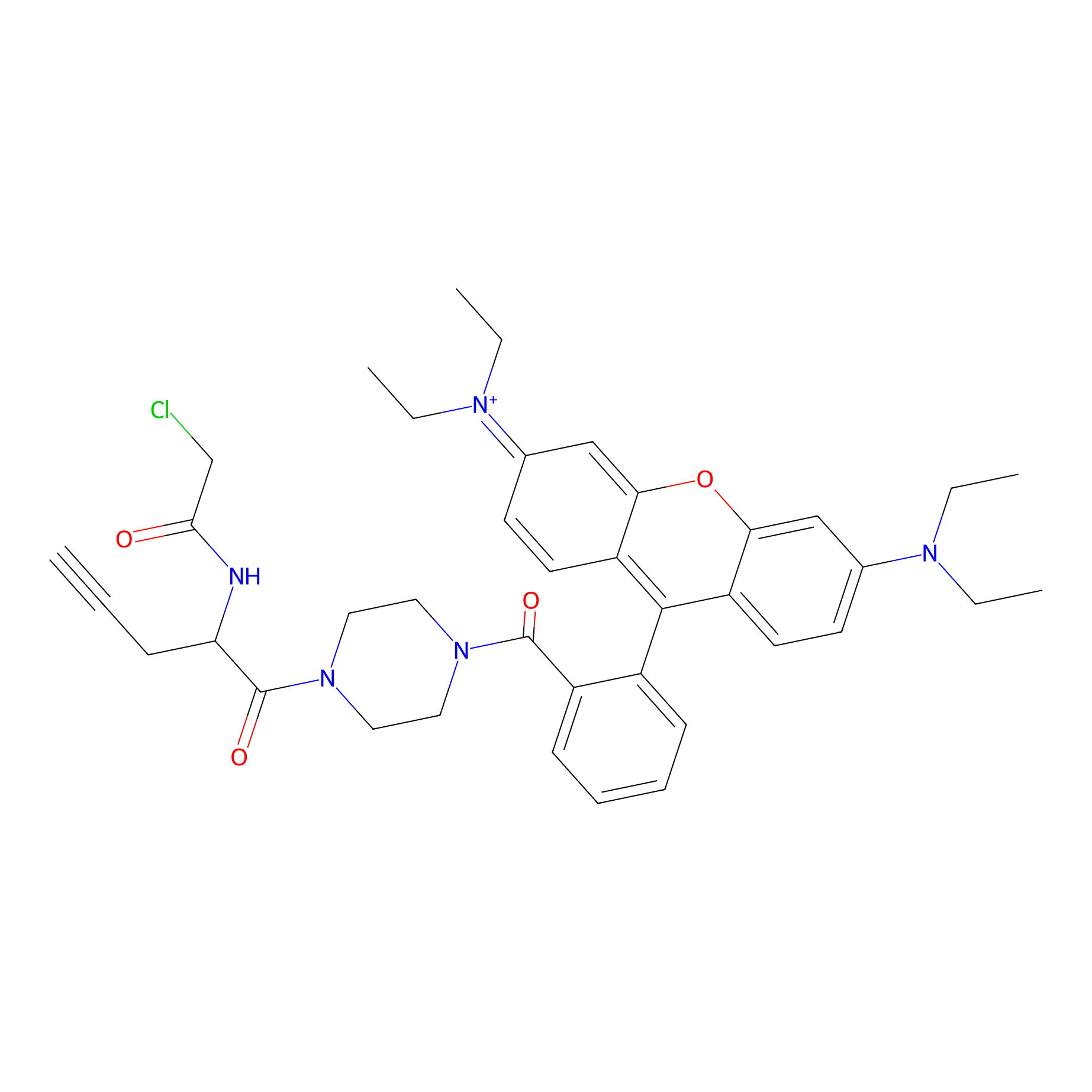 |
N.A. | LDD0426 | [14] | |
|
TPP-AC Probe Info |
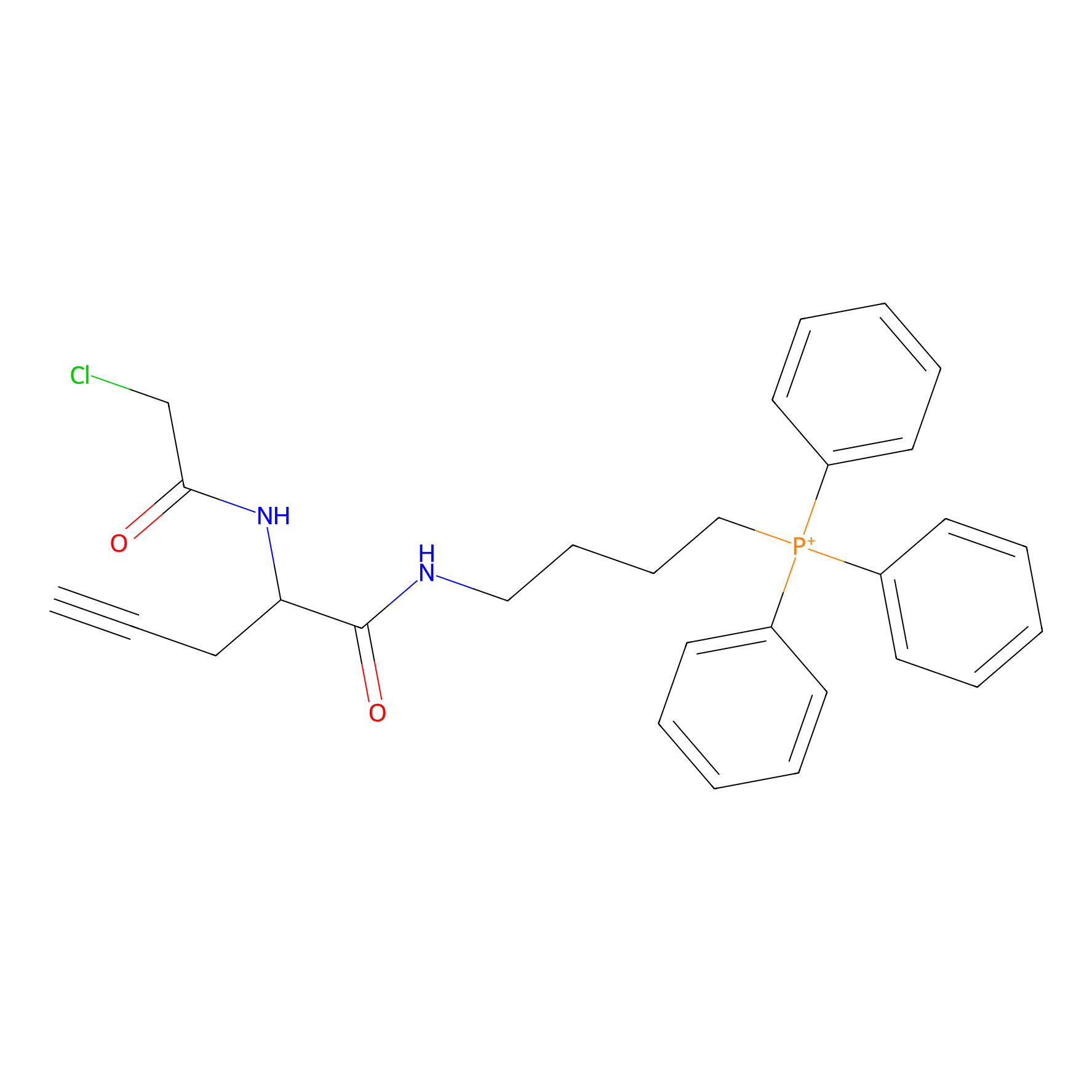 |
N.A. | LDD0427 | [14] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe11 Probe Info |
 |
20.00 | LDD0471 | [15] | |
|
FFF probe14 Probe Info |
 |
20.00 | LDD0477 | [15] | |
|
FFF probe2 Probe Info |
 |
18.94 | LDD0463 | [15] | |
|
FFF probe3 Probe Info |
 |
13.81 | LDD0464 | [15] | |
|
JN0003 Probe Info |
 |
20.00 | LDD0469 | [15] | |
|
OEA-DA Probe Info |
 |
3.09 | LDD0046 | [16] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HEK-293T | C183(1.00) | LDD1507 | [17] |
| LDCM0226 | AC11 | HEK-293T | C183(0.90) | LDD1509 | [17] |
| LDCM0259 | AC14 | HEK-293T | C183(1.01) | LDD1512 | [17] |
| LDCM0270 | AC15 | HEK-293T | C183(0.91); C88(0.85) | LDD1513 | [17] |
| LDCM0276 | AC17 | HEK-293T | C183(1.14) | LDD1515 | [17] |
| LDCM0278 | AC19 | HEK-293T | C183(0.88) | LDD1517 | [17] |
| LDCM0281 | AC21 | HEK-293T | C183(0.87) | LDD1520 | [17] |
| LDCM0282 | AC22 | HEK-293T | C183(1.04) | LDD1521 | [17] |
| LDCM0283 | AC23 | HEK-293T | C183(0.99); C88(0.75) | LDD1522 | [17] |
| LDCM0284 | AC24 | HEK-293T | C183(0.97) | LDD1523 | [17] |
| LDCM0285 | AC25 | HEK-293T | C183(0.97) | LDD1524 | [17] |
| LDCM0287 | AC27 | HEK-293T | C183(0.82) | LDD1526 | [17] |
| LDCM0289 | AC29 | HEK-293T | C183(0.89) | LDD1528 | [17] |
| LDCM0290 | AC3 | HEK-293T | C183(0.98) | LDD1529 | [17] |
| LDCM0291 | AC30 | HEK-293T | C183(0.80) | LDD1530 | [17] |
| LDCM0292 | AC31 | HEK-293T | C183(0.95); C88(0.87) | LDD1531 | [17] |
| LDCM0293 | AC32 | HEK-293T | C183(0.96) | LDD1532 | [17] |
| LDCM0294 | AC33 | HEK-293T | C183(1.03) | LDD1533 | [17] |
| LDCM0296 | AC35 | HEK-293T | C183(0.92) | LDD1535 | [17] |
| LDCM0298 | AC37 | HEK-293T | C183(0.91) | LDD1537 | [17] |
| LDCM0299 | AC38 | HEK-293T | C183(0.93) | LDD1538 | [17] |
| LDCM0300 | AC39 | HEK-293T | C183(0.99); C88(0.90) | LDD1539 | [17] |
| LDCM0302 | AC40 | HEK-293T | C183(1.21) | LDD1541 | [17] |
| LDCM0303 | AC41 | HEK-293T | C183(1.01) | LDD1542 | [17] |
| LDCM0305 | AC43 | HEK-293T | C183(0.96) | LDD1544 | [17] |
| LDCM0307 | AC45 | HEK-293T | C183(0.82) | LDD1546 | [17] |
| LDCM0308 | AC46 | HEK-293T | C183(1.12) | LDD1547 | [17] |
| LDCM0309 | AC47 | HEK-293T | C183(0.97); C88(1.08) | LDD1548 | [17] |
| LDCM0310 | AC48 | HEK-293T | C183(1.28) | LDD1549 | [17] |
| LDCM0311 | AC49 | HEK-293T | C183(0.93) | LDD1550 | [17] |
| LDCM0312 | AC5 | HEK-293T | C183(0.91) | LDD1551 | [17] |
| LDCM0314 | AC51 | HEK-293T | C183(0.92) | LDD1553 | [17] |
| LDCM0316 | AC53 | HEK-293T | C183(0.80) | LDD1555 | [17] |
| LDCM0317 | AC54 | HEK-293T | C183(1.35) | LDD1556 | [17] |
| LDCM0318 | AC55 | HEK-293T | C183(0.95); C88(0.91) | LDD1557 | [17] |
| LDCM0319 | AC56 | HEK-293T | C183(0.83) | LDD1558 | [17] |
| LDCM0320 | AC57 | HEK-293T | C183(1.48) | LDD1559 | [17] |
| LDCM0322 | AC59 | HEK-293T | C183(1.01) | LDD1561 | [17] |
| LDCM0323 | AC6 | HEK-293T | C183(1.01) | LDD1562 | [17] |
| LDCM0325 | AC61 | HEK-293T | C183(0.98) | LDD1564 | [17] |
| LDCM0326 | AC62 | HEK-293T | C183(0.94) | LDD1565 | [17] |
| LDCM0327 | AC63 | HEK-293T | C183(1.10); C88(0.87) | LDD1566 | [17] |
| LDCM0328 | AC64 | HEK-293T | C183(1.24) | LDD1567 | [17] |
| LDCM0334 | AC7 | HEK-293T | C183(1.06); C88(0.87) | LDD1568 | [17] |
| LDCM0345 | AC8 | HEK-293T | C183(1.03) | LDD1569 | [17] |
| LDCM0248 | AKOS034007472 | HEK-293T | C183(0.99) | LDD1511 | [17] |
| LDCM0356 | AKOS034007680 | HEK-293T | C183(0.87) | LDD1570 | [17] |
| LDCM0275 | AKOS034007705 | HEK-293T | C183(1.00) | LDD1514 | [17] |
| LDCM0630 | CCW28-3 | 231MFP | C183(1.14) | LDD2214 | [18] |
| LDCM0367 | CL1 | HEK-293T | C183(1.30) | LDD1571 | [17] |
| LDCM0368 | CL10 | HEK-293T | C183(0.91) | LDD1572 | [17] |
| LDCM0369 | CL100 | HEK-293T | C183(0.93) | LDD1573 | [17] |
| LDCM0370 | CL101 | HEK-293T | C183(1.07) | LDD1574 | [17] |
| LDCM0371 | CL102 | HEK-293T | C183(1.29) | LDD1575 | [17] |
| LDCM0372 | CL103 | HEK-293T | C183(1.65) | LDD1576 | [17] |
| LDCM0373 | CL104 | HEK-293T | C183(1.00) | LDD1577 | [17] |
| LDCM0374 | CL105 | HEK-293T | C183(0.93) | LDD1578 | [17] |
| LDCM0375 | CL106 | HEK-293T | C183(0.95) | LDD1579 | [17] |
| LDCM0376 | CL107 | HEK-293T | C183(0.95) | LDD1580 | [17] |
| LDCM0377 | CL108 | HEK-293T | C183(1.13) | LDD1581 | [17] |
| LDCM0378 | CL109 | HEK-293T | C183(0.96) | LDD1582 | [17] |
| LDCM0379 | CL11 | HEK-293T | C183(1.21); C88(0.64) | LDD1583 | [17] |
| LDCM0380 | CL110 | HEK-293T | C183(1.01) | LDD1584 | [17] |
| LDCM0381 | CL111 | HEK-293T | C183(0.63) | LDD1585 | [17] |
| LDCM0382 | CL112 | HEK-293T | C183(1.04) | LDD1586 | [17] |
| LDCM0383 | CL113 | HEK-293T | C183(1.35) | LDD1587 | [17] |
| LDCM0384 | CL114 | HEK-293T | C183(0.90) | LDD1588 | [17] |
| LDCM0385 | CL115 | HEK-293T | C183(0.78) | LDD1589 | [17] |
| LDCM0386 | CL116 | HEK-293T | C183(1.02) | LDD1590 | [17] |
| LDCM0387 | CL117 | HEK-293T | C183(1.10) | LDD1591 | [17] |
| LDCM0388 | CL118 | HEK-293T | C183(0.84) | LDD1592 | [17] |
| LDCM0389 | CL119 | HEK-293T | C183(0.90) | LDD1593 | [17] |
| LDCM0390 | CL12 | HEK-293T | C183(1.31) | LDD1594 | [17] |
| LDCM0391 | CL120 | HEK-293T | C183(1.04) | LDD1595 | [17] |
| LDCM0392 | CL121 | HEK-293T | C183(0.94) | LDD1596 | [17] |
| LDCM0393 | CL122 | HEK-293T | C183(1.42) | LDD1597 | [17] |
| LDCM0394 | CL123 | HEK-293T | C183(1.11) | LDD1598 | [17] |
| LDCM0395 | CL124 | HEK-293T | C183(0.97) | LDD1599 | [17] |
| LDCM0396 | CL125 | HEK-293T | C183(1.05) | LDD1600 | [17] |
| LDCM0397 | CL126 | HEK-293T | C183(1.07) | LDD1601 | [17] |
| LDCM0398 | CL127 | HEK-293T | C183(0.98) | LDD1602 | [17] |
| LDCM0399 | CL128 | HEK-293T | C183(1.24) | LDD1603 | [17] |
| LDCM0400 | CL13 | HEK-293T | C183(0.95) | LDD1604 | [17] |
| LDCM0401 | CL14 | HEK-293T | C183(1.33) | LDD1605 | [17] |
| LDCM0402 | CL15 | HEK-293T | C183(2.04) | LDD1606 | [17] |
| LDCM0403 | CL16 | HEK-293T | C183(1.67) | LDD1607 | [17] |
| LDCM0404 | CL17 | HEK-293T | C183(2.69) | LDD1608 | [17] |
| LDCM0406 | CL19 | HEK-293T | C183(1.07) | LDD1610 | [17] |
| LDCM0407 | CL2 | HEK-293T | C183(1.47) | LDD1611 | [17] |
| LDCM0409 | CL21 | HEK-293T | C183(0.57) | LDD1613 | [17] |
| LDCM0410 | CL22 | HEK-293T | C183(1.12) | LDD1614 | [17] |
| LDCM0411 | CL23 | HEK-293T | C183(0.94); C88(0.76) | LDD1615 | [17] |
| LDCM0412 | CL24 | HEK-293T | C183(1.23) | LDD1616 | [17] |
| LDCM0413 | CL25 | HEK-293T | C183(0.99) | LDD1617 | [17] |
| LDCM0414 | CL26 | HEK-293T | C183(1.65) | LDD1618 | [17] |
| LDCM0415 | CL27 | HEK-293T | C183(0.75) | LDD1619 | [17] |
| LDCM0416 | CL28 | HEK-293T | C183(1.21) | LDD1620 | [17] |
| LDCM0417 | CL29 | HEK-293T | C183(0.96) | LDD1621 | [17] |
| LDCM0418 | CL3 | HEK-293T | C183(1.40) | LDD1622 | [17] |
| LDCM0420 | CL31 | HEK-293T | C183(0.85) | LDD1624 | [17] |
| LDCM0422 | CL33 | HEK-293T | C183(0.95) | LDD1626 | [17] |
| LDCM0423 | CL34 | HEK-293T | C183(0.97) | LDD1627 | [17] |
| LDCM0424 | CL35 | HEK-293T | C183(1.47); C88(0.61) | LDD1628 | [17] |
| LDCM0425 | CL36 | HEK-293T | C183(1.18) | LDD1629 | [17] |
| LDCM0426 | CL37 | HEK-293T | C183(1.14) | LDD1630 | [17] |
| LDCM0428 | CL39 | HEK-293T | C183(1.03) | LDD1632 | [17] |
| LDCM0429 | CL4 | HEK-293T | C183(1.00) | LDD1633 | [17] |
| LDCM0430 | CL40 | HEK-293T | C183(1.02) | LDD1634 | [17] |
| LDCM0431 | CL41 | HEK-293T | C183(0.79) | LDD1635 | [17] |
| LDCM0433 | CL43 | HEK-293T | C183(0.87) | LDD1637 | [17] |
| LDCM0435 | CL45 | HEK-293T | C183(1.05) | LDD1639 | [17] |
| LDCM0436 | CL46 | HEK-293T | C183(0.91) | LDD1640 | [17] |
| LDCM0437 | CL47 | HEK-293T | C183(1.15); C88(0.74) | LDD1641 | [17] |
| LDCM0438 | CL48 | HEK-293T | C183(1.40) | LDD1642 | [17] |
| LDCM0439 | CL49 | HEK-293T | C183(1.22) | LDD1643 | [17] |
| LDCM0440 | CL5 | HEK-293T | C183(1.67) | LDD1644 | [17] |
| LDCM0441 | CL50 | HEK-293T | C183(1.44) | LDD1645 | [17] |
| LDCM0443 | CL52 | HEK-293T | C183(0.97) | LDD1646 | [17] |
| LDCM0444 | CL53 | HEK-293T | C183(1.23) | LDD1647 | [17] |
| LDCM0446 | CL55 | HEK-293T | C183(1.04) | LDD1649 | [17] |
| LDCM0448 | CL57 | HEK-293T | C183(0.79) | LDD1651 | [17] |
| LDCM0449 | CL58 | HEK-293T | C183(1.16) | LDD1652 | [17] |
| LDCM0450 | CL59 | HEK-293T | C183(1.00); C88(0.81) | LDD1653 | [17] |
| LDCM0452 | CL60 | HEK-293T | C183(1.34) | LDD1655 | [17] |
| LDCM0453 | CL61 | HEK-293T | C183(1.16) | LDD1656 | [17] |
| LDCM0454 | CL62 | HEK-293T | C183(1.31) | LDD1657 | [17] |
| LDCM0455 | CL63 | HEK-293T | C183(1.25) | LDD1658 | [17] |
| LDCM0456 | CL64 | HEK-293T | C183(1.34) | LDD1659 | [17] |
| LDCM0457 | CL65 | HEK-293T | C183(1.14) | LDD1660 | [17] |
| LDCM0459 | CL67 | HEK-293T | C183(1.06) | LDD1662 | [17] |
| LDCM0461 | CL69 | HEK-293T | C183(1.06) | LDD1664 | [17] |
| LDCM0462 | CL7 | HEK-293T | C183(0.97) | LDD1665 | [17] |
| LDCM0463 | CL70 | HEK-293T | C183(1.09) | LDD1666 | [17] |
| LDCM0464 | CL71 | HEK-293T | C183(0.97); C88(0.65) | LDD1667 | [17] |
| LDCM0465 | CL72 | HEK-293T | C183(1.51) | LDD1668 | [17] |
| LDCM0466 | CL73 | HEK-293T | C183(1.09) | LDD1669 | [17] |
| LDCM0467 | CL74 | HEK-293T | C183(1.05) | LDD1670 | [17] |
| LDCM0469 | CL76 | HEK-293T | C183(1.15) | LDD1672 | [17] |
| LDCM0470 | CL77 | HEK-293T | C183(1.06) | LDD1673 | [17] |
| LDCM0472 | CL79 | HEK-293T | C183(0.86) | LDD1675 | [17] |
| LDCM0475 | CL81 | HEK-293T | C183(1.05) | LDD1678 | [17] |
| LDCM0476 | CL82 | HEK-293T | C183(1.00) | LDD1679 | [17] |
| LDCM0477 | CL83 | HEK-293T | C183(1.09); C88(0.72) | LDD1680 | [17] |
| LDCM0478 | CL84 | HEK-293T | C183(1.16) | LDD1681 | [17] |
| LDCM0479 | CL85 | HEK-293T | C183(0.97) | LDD1682 | [17] |
| LDCM0480 | CL86 | HEK-293T | C183(0.90) | LDD1683 | [17] |
| LDCM0481 | CL87 | HEK-293T | C183(0.83) | LDD1684 | [17] |
| LDCM0482 | CL88 | HEK-293T | C183(1.29) | LDD1685 | [17] |
| LDCM0483 | CL89 | HEK-293T | C183(1.10) | LDD1686 | [17] |
| LDCM0484 | CL9 | HEK-293T | C183(0.98) | LDD1687 | [17] |
| LDCM0486 | CL91 | HEK-293T | C183(0.94) | LDD1689 | [17] |
| LDCM0488 | CL93 | HEK-293T | C183(0.86) | LDD1691 | [17] |
| LDCM0489 | CL94 | HEK-293T | C183(0.96) | LDD1692 | [17] |
| LDCM0490 | CL95 | HEK-293T | C183(1.15); C88(0.93) | LDD1693 | [17] |
| LDCM0491 | CL96 | HEK-293T | C183(1.24) | LDD1694 | [17] |
| LDCM0492 | CL97 | HEK-293T | C183(1.27) | LDD1695 | [17] |
| LDCM0493 | CL98 | HEK-293T | C183(1.59) | LDD1696 | [17] |
| LDCM0494 | CL99 | HEK-293T | C183(0.92) | LDD1697 | [17] |
| LDCM0495 | E2913 | HEK-293T | C183(1.07) | LDD1698 | [17] |
| LDCM0157 | EN6 | HEK293-A | C194(4.56) | LDD0406 | [6] |
| LDCM0468 | Fragment33 | HEK-293T | C183(1.15) | LDD1671 | [17] |
| LDCM0427 | Fragment51 | HEK-293T | C183(0.67) | LDD1631 | [17] |
| LDCM0022 | KB02 | AN3-CA | C183(1.05) | LDD2264 | [4] |
| LDCM0023 | KB03 | AN3-CA | C183(0.97) | LDD2681 | [4] |
| LDCM0024 | KB05 | NCI-H1155 | C183(1.25) | LDD3342 | [4] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0224 | [12] |
| LDCM0016 | Ranjitkar_cp1 | MDA-MB-231 | 5.88 | LDD0123 | [5] |
The Interaction Atlas With This Target
References
