Details of the Target
General Information of Target
| Target ID | LDTP01057 | |||||
|---|---|---|---|---|---|---|
| Target Name | Polyglutamine-binding protein 1 (PQBP1) | |||||
| Gene Name | PQBP1 | |||||
| Gene ID | 10084 | |||||
| Synonyms |
NPW38; Polyglutamine-binding protein 1; PQBP-1; 38 kDa nuclear protein containing a WW domain; Npw38; Polyglutamine tract-binding protein 1 |
|||||
| 3D Structure | ||||||
| Sequence |
MPLPVALQTRLAKRGILKHLEPEPEEEIIAEDYDDDPVDYEATRLEGLPPSWYKVFDPSC
GLPYYWNADTDLVSWLSPHDPNSVVTKSAKKLRSSNADAEEKLDRSHDKSDRGHDKSDRS HEKLDRGHDKSDRGHDKSDRDRERGYDKVDRERERDRERDRDRGYDKADREEGKERRHHR REELAPYPKSKKAVSRKDEELDPMDPSSYSDAPRGTWSTGLPKRNEAKTGADTTAAGPLF QQRPYPSPGAVLRANAEASRTKQQD |
|||||
| Target Bioclass |
Other
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Intrinsically disordered protein that acts as a scaffold, and which is involved in different processes, such as pre-mRNA splicing, transcription regulation, innate immunity and neuron development. Interacts with splicing-related factors via the intrinsically disordered region and regulates alternative splicing of target pre-mRNA species. May suppress the ability of POU3F2 to transactivate the DRD1 gene in a POU3F2 dependent manner. Can activate transcription directly or via association with the transcription machinery. May be involved in ATXN1 mutant-induced cell death. The interaction with ATXN1 mutant reduces levels of phosphorylated RNA polymerase II large subunit. Involved in the assembly of cytoplasmic stress granule, possibly by participating in the transport of neuronal RNA granules. Also acts as an innate immune sensor of infection by retroviruses, such as HIV, by detecting the presence of reverse-transcribed DNA in the cytosol. Directly binds retroviral reverse-transcribed DNA in the cytosol and interacts with CGAS, leading to activate the cGAS-STING signaling pathway, triggering type-I interferon production.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
13.24 | LDD0402 | [1] | |
|
FBP2 Probe Info |
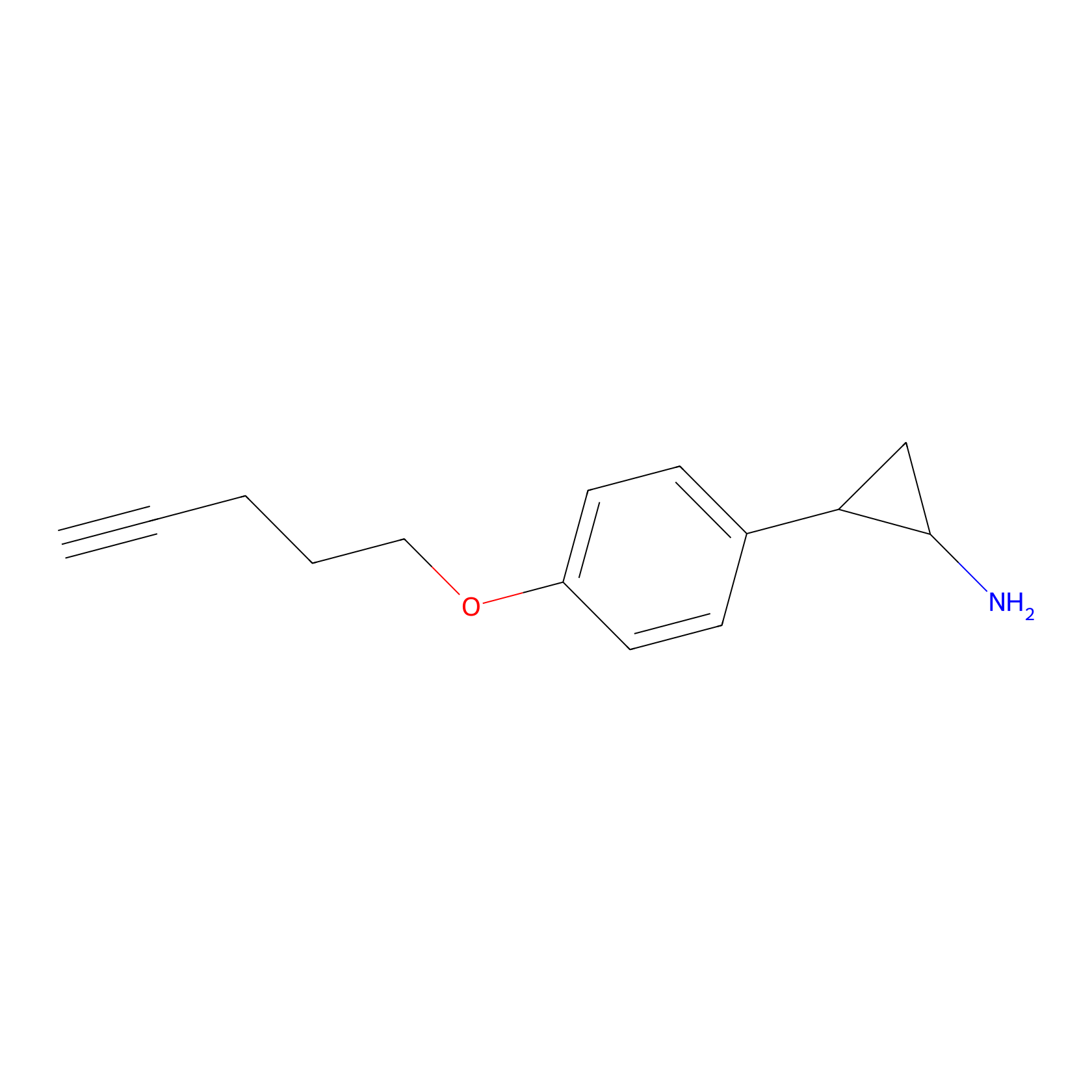 |
8.81 | LDD0323 | [2] | |
|
STPyne Probe Info |
 |
K123(5.26) | LDD0277 | [3] | |
|
Probe 1 Probe Info |
 |
Y33(27.24); Y40(24.87); Y209(5.57); Y245(11.11) | LDD3495 | [4] | |
|
DBIA Probe Info |
 |
C60(0.84) | LDD3312 | [5] | |
|
AHL-Pu-1 Probe Info |
 |
C60(5.87) | LDD0169 | [6] | |
|
HHS-475 Probe Info |
 |
Y245(0.79); Y209(6.96) | LDD0264 | [7] | |
|
HHS-465 Probe Info |
 |
Y187(4.83); Y245(1.25) | LDD2237 | [8] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0221 | [9] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [10] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [10] | |
|
IPIAA_H Probe Info |
 |
N.A. | LDD0030 | [11] | |
|
IPIAA_L Probe Info |
 |
N.A. | LDD0031 | [11] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [10] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [12] | |
|
IPM Probe Info |
 |
N.A. | LDD2156 | [13] | |
|
SF Probe Info |
 |
Y245(0.00); Y146(0.00); Y187(0.00) | LDD0028 | [14] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [15] | |
|
AOyne Probe Info |
 |
12.80 | LDD0443 | [16] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [17] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C60(5.87) | LDD0169 | [6] |
| LDCM0214 | AC1 | HEK-293T | C60(1.06) | LDD1507 | [18] |
| LDCM0215 | AC10 | HEK-293T | C60(0.88) | LDD1508 | [18] |
| LDCM0226 | AC11 | HEK-293T | C60(0.95) | LDD1509 | [18] |
| LDCM0237 | AC12 | HEK-293T | C60(0.93) | LDD1510 | [18] |
| LDCM0259 | AC14 | HEK-293T | C60(1.07) | LDD1512 | [18] |
| LDCM0270 | AC15 | HEK-293T | C60(1.12) | LDD1513 | [18] |
| LDCM0276 | AC17 | HEK-293T | C60(0.96) | LDD1515 | [18] |
| LDCM0277 | AC18 | HEK-293T | C60(0.91) | LDD1516 | [18] |
| LDCM0278 | AC19 | HEK-293T | C60(0.82) | LDD1517 | [18] |
| LDCM0279 | AC2 | HEK-293T | C60(0.99) | LDD1518 | [18] |
| LDCM0280 | AC20 | HEK-293T | C60(0.96) | LDD1519 | [18] |
| LDCM0281 | AC21 | HEK-293T | C60(1.01) | LDD1520 | [18] |
| LDCM0282 | AC22 | HEK-293T | C60(1.09) | LDD1521 | [18] |
| LDCM0283 | AC23 | HEK-293T | C60(1.12) | LDD1522 | [18] |
| LDCM0284 | AC24 | HEK-293T | C60(1.04) | LDD1523 | [18] |
| LDCM0285 | AC25 | HEK-293T | C60(0.96) | LDD1524 | [18] |
| LDCM0286 | AC26 | HEK-293T | C60(0.95) | LDD1525 | [18] |
| LDCM0287 | AC27 | HEK-293T | C60(1.08) | LDD1526 | [18] |
| LDCM0288 | AC28 | HEK-293T | C60(0.87) | LDD1527 | [18] |
| LDCM0289 | AC29 | HEK-293T | C60(0.95) | LDD1528 | [18] |
| LDCM0290 | AC3 | HEK-293T | C60(0.92) | LDD1529 | [18] |
| LDCM0291 | AC30 | HEK-293T | C60(1.06) | LDD1530 | [18] |
| LDCM0292 | AC31 | HEK-293T | C60(1.10) | LDD1531 | [18] |
| LDCM0293 | AC32 | HEK-293T | C60(0.99) | LDD1532 | [18] |
| LDCM0294 | AC33 | HEK-293T | C60(0.96) | LDD1533 | [18] |
| LDCM0295 | AC34 | HEK-293T | C60(0.96) | LDD1534 | [18] |
| LDCM0296 | AC35 | HEK-293T | C60(1.01) | LDD1535 | [18] |
| LDCM0297 | AC36 | HEK-293T | C60(0.91) | LDD1536 | [18] |
| LDCM0298 | AC37 | HEK-293T | C60(1.05) | LDD1537 | [18] |
| LDCM0299 | AC38 | HEK-293T | C60(0.95) | LDD1538 | [18] |
| LDCM0300 | AC39 | HEK-293T | C60(1.11) | LDD1539 | [18] |
| LDCM0301 | AC4 | HEK-293T | C60(0.93) | LDD1540 | [18] |
| LDCM0302 | AC40 | HEK-293T | C60(1.16) | LDD1541 | [18] |
| LDCM0303 | AC41 | HEK-293T | C60(0.97) | LDD1542 | [18] |
| LDCM0304 | AC42 | HEK-293T | C60(1.04) | LDD1543 | [18] |
| LDCM0305 | AC43 | HEK-293T | C60(1.06) | LDD1544 | [18] |
| LDCM0306 | AC44 | HEK-293T | C60(0.91) | LDD1545 | [18] |
| LDCM0307 | AC45 | HEK-293T | C60(0.93) | LDD1546 | [18] |
| LDCM0308 | AC46 | HEK-293T | C60(1.12) | LDD1547 | [18] |
| LDCM0309 | AC47 | HEK-293T | C60(1.01) | LDD1548 | [18] |
| LDCM0310 | AC48 | HEK-293T | C60(1.10) | LDD1549 | [18] |
| LDCM0311 | AC49 | HEK-293T | C60(1.02) | LDD1550 | [18] |
| LDCM0312 | AC5 | HEK-293T | C60(0.93) | LDD1551 | [18] |
| LDCM0313 | AC50 | HEK-293T | C60(0.87) | LDD1552 | [18] |
| LDCM0314 | AC51 | HEK-293T | C60(1.03) | LDD1553 | [18] |
| LDCM0315 | AC52 | HEK-293T | C60(0.85) | LDD1554 | [18] |
| LDCM0316 | AC53 | HEK-293T | C60(1.02) | LDD1555 | [18] |
| LDCM0317 | AC54 | HEK-293T | C60(0.92) | LDD1556 | [18] |
| LDCM0318 | AC55 | HEK-293T | C60(1.07) | LDD1557 | [18] |
| LDCM0319 | AC56 | HEK-293T | C60(0.95) | LDD1558 | [18] |
| LDCM0320 | AC57 | HEK-293T | C60(0.95) | LDD1559 | [18] |
| LDCM0321 | AC58 | HEK-293T | C60(0.91) | LDD1560 | [18] |
| LDCM0322 | AC59 | HEK-293T | C60(1.09) | LDD1561 | [18] |
| LDCM0323 | AC6 | HEK-293T | C60(1.01) | LDD1562 | [18] |
| LDCM0324 | AC60 | HEK-293T | C60(0.91) | LDD1563 | [18] |
| LDCM0325 | AC61 | HEK-293T | C60(0.87) | LDD1564 | [18] |
| LDCM0326 | AC62 | HEK-293T | C60(1.08) | LDD1565 | [18] |
| LDCM0327 | AC63 | HEK-293T | C60(1.03) | LDD1566 | [18] |
| LDCM0328 | AC64 | HEK-293T | C60(0.98) | LDD1567 | [18] |
| LDCM0334 | AC7 | HEK-293T | C60(1.27) | LDD1568 | [18] |
| LDCM0345 | AC8 | HEK-293T | C60(1.07) | LDD1569 | [18] |
| LDCM0248 | AKOS034007472 | HEK-293T | C60(0.97) | LDD1511 | [18] |
| LDCM0356 | AKOS034007680 | HEK-293T | C60(1.04) | LDD1570 | [18] |
| LDCM0275 | AKOS034007705 | HEK-293T | C60(1.06) | LDD1514 | [18] |
| LDCM0156 | Aniline | NCI-H1299 | 12.81 | LDD0403 | [1] |
| LDCM0630 | CCW28-3 | 231MFP | C60(1.77) | LDD2214 | [19] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [9] |
| LDCM0367 | CL1 | HEK-293T | C60(0.97) | LDD1571 | [18] |
| LDCM0368 | CL10 | HEK-293T | C60(0.90) | LDD1572 | [18] |
| LDCM0369 | CL100 | HEK-293T | C60(0.83) | LDD1573 | [18] |
| LDCM0370 | CL101 | HEK-293T | C60(0.93) | LDD1574 | [18] |
| LDCM0371 | CL102 | HEK-293T | C60(0.95) | LDD1575 | [18] |
| LDCM0372 | CL103 | HEK-293T | C60(0.93) | LDD1576 | [18] |
| LDCM0373 | CL104 | HEK-293T | C60(0.82) | LDD1577 | [18] |
| LDCM0374 | CL105 | HEK-293T | C60(0.97) | LDD1578 | [18] |
| LDCM0375 | CL106 | HEK-293T | C60(0.94) | LDD1579 | [18] |
| LDCM0376 | CL107 | HEK-293T | C60(0.95) | LDD1580 | [18] |
| LDCM0377 | CL108 | HEK-293T | C60(0.91) | LDD1581 | [18] |
| LDCM0378 | CL109 | HEK-293T | C60(0.94) | LDD1582 | [18] |
| LDCM0379 | CL11 | HEK-293T | C60(0.93) | LDD1583 | [18] |
| LDCM0380 | CL110 | HEK-293T | C60(0.93) | LDD1584 | [18] |
| LDCM0381 | CL111 | HEK-293T | C60(0.99) | LDD1585 | [18] |
| LDCM0382 | CL112 | HEK-293T | C60(1.11) | LDD1586 | [18] |
| LDCM0383 | CL113 | HEK-293T | C60(1.04) | LDD1587 | [18] |
| LDCM0384 | CL114 | HEK-293T | C60(0.97) | LDD1588 | [18] |
| LDCM0385 | CL115 | HEK-293T | C60(0.97) | LDD1589 | [18] |
| LDCM0386 | CL116 | HEK-293T | C60(0.96) | LDD1590 | [18] |
| LDCM0387 | CL117 | HEK-293T | C60(0.93) | LDD1591 | [18] |
| LDCM0388 | CL118 | HEK-293T | C60(1.05) | LDD1592 | [18] |
| LDCM0389 | CL119 | HEK-293T | C60(0.99) | LDD1593 | [18] |
| LDCM0390 | CL12 | HEK-293T | C60(0.98) | LDD1594 | [18] |
| LDCM0391 | CL120 | HEK-293T | C60(0.91) | LDD1595 | [18] |
| LDCM0392 | CL121 | HEK-293T | C60(0.93) | LDD1596 | [18] |
| LDCM0393 | CL122 | HEK-293T | C60(0.97) | LDD1597 | [18] |
| LDCM0394 | CL123 | HEK-293T | C60(0.79) | LDD1598 | [18] |
| LDCM0395 | CL124 | HEK-293T | C60(0.85) | LDD1599 | [18] |
| LDCM0396 | CL125 | HEK-293T | C60(0.79) | LDD1600 | [18] |
| LDCM0397 | CL126 | HEK-293T | C60(0.88) | LDD1601 | [18] |
| LDCM0398 | CL127 | HEK-293T | C60(0.86) | LDD1602 | [18] |
| LDCM0399 | CL128 | HEK-293T | C60(0.79) | LDD1603 | [18] |
| LDCM0400 | CL13 | HEK-293T | C60(0.93) | LDD1604 | [18] |
| LDCM0401 | CL14 | HEK-293T | C60(1.11) | LDD1605 | [18] |
| LDCM0402 | CL15 | HEK-293T | C60(0.91) | LDD1606 | [18] |
| LDCM0403 | CL16 | HEK-293T | C60(0.96) | LDD1607 | [18] |
| LDCM0404 | CL17 | HEK-293T | C60(0.98) | LDD1608 | [18] |
| LDCM0405 | CL18 | HEK-293T | C60(0.87) | LDD1609 | [18] |
| LDCM0406 | CL19 | HEK-293T | C60(0.98) | LDD1610 | [18] |
| LDCM0407 | CL2 | HEK-293T | C60(1.13) | LDD1611 | [18] |
| LDCM0408 | CL20 | HEK-293T | C60(0.91) | LDD1612 | [18] |
| LDCM0409 | CL21 | HEK-293T | C60(0.95) | LDD1613 | [18] |
| LDCM0410 | CL22 | HEK-293T | C60(1.04) | LDD1614 | [18] |
| LDCM0411 | CL23 | HEK-293T | C60(1.11) | LDD1615 | [18] |
| LDCM0412 | CL24 | HEK-293T | C60(1.02) | LDD1616 | [18] |
| LDCM0413 | CL25 | HEK-293T | C60(0.80) | LDD1617 | [18] |
| LDCM0414 | CL26 | HEK-293T | C60(1.10) | LDD1618 | [18] |
| LDCM0415 | CL27 | HEK-293T | C60(1.04) | LDD1619 | [18] |
| LDCM0416 | CL28 | HEK-293T | C60(0.95) | LDD1620 | [18] |
| LDCM0417 | CL29 | HEK-293T | C60(0.92) | LDD1621 | [18] |
| LDCM0418 | CL3 | HEK-293T | C60(0.84) | LDD1622 | [18] |
| LDCM0419 | CL30 | HEK-293T | C60(0.94) | LDD1623 | [18] |
| LDCM0420 | CL31 | HEK-293T | C60(0.99) | LDD1624 | [18] |
| LDCM0421 | CL32 | HEK-293T | C60(0.81) | LDD1625 | [18] |
| LDCM0422 | CL33 | HEK-293T | C60(0.84) | LDD1626 | [18] |
| LDCM0423 | CL34 | HEK-293T | C60(0.86) | LDD1627 | [18] |
| LDCM0424 | CL35 | HEK-293T | C60(1.13) | LDD1628 | [18] |
| LDCM0425 | CL36 | HEK-293T | C60(0.98) | LDD1629 | [18] |
| LDCM0426 | CL37 | HEK-293T | C60(1.02) | LDD1630 | [18] |
| LDCM0428 | CL39 | HEK-293T | C60(1.00) | LDD1632 | [18] |
| LDCM0429 | CL4 | HEK-293T | C60(0.76) | LDD1633 | [18] |
| LDCM0430 | CL40 | HEK-293T | C60(0.93) | LDD1634 | [18] |
| LDCM0431 | CL41 | HEK-293T | C60(0.98) | LDD1635 | [18] |
| LDCM0432 | CL42 | HEK-293T | C60(0.84) | LDD1636 | [18] |
| LDCM0433 | CL43 | HEK-293T | C60(1.04) | LDD1637 | [18] |
| LDCM0434 | CL44 | HEK-293T | C60(0.73) | LDD1638 | [18] |
| LDCM0435 | CL45 | HEK-293T | C60(0.85) | LDD1639 | [18] |
| LDCM0436 | CL46 | HEK-293T | C60(0.96) | LDD1640 | [18] |
| LDCM0437 | CL47 | HEK-293T | C60(1.12) | LDD1641 | [18] |
| LDCM0438 | CL48 | HEK-293T | C60(0.97) | LDD1642 | [18] |
| LDCM0439 | CL49 | HEK-293T | C60(0.93) | LDD1643 | [18] |
| LDCM0440 | CL5 | HEK-293T | C60(1.03) | LDD1644 | [18] |
| LDCM0441 | CL50 | HEK-293T | C60(0.99) | LDD1645 | [18] |
| LDCM0443 | CL52 | HEK-293T | C60(0.81) | LDD1646 | [18] |
| LDCM0444 | CL53 | HEK-293T | C60(0.86) | LDD1647 | [18] |
| LDCM0445 | CL54 | HEK-293T | C60(0.82) | LDD1648 | [18] |
| LDCM0446 | CL55 | HEK-293T | C60(0.86) | LDD1649 | [18] |
| LDCM0447 | CL56 | HEK-293T | C60(0.79) | LDD1650 | [18] |
| LDCM0448 | CL57 | HEK-293T | C60(0.88) | LDD1651 | [18] |
| LDCM0449 | CL58 | HEK-293T | C60(0.94) | LDD1652 | [18] |
| LDCM0450 | CL59 | HEK-293T | C60(0.94) | LDD1653 | [18] |
| LDCM0451 | CL6 | HEK-293T | C60(0.86) | LDD1654 | [18] |
| LDCM0452 | CL60 | HEK-293T | C60(0.97) | LDD1655 | [18] |
| LDCM0453 | CL61 | HEK-293T | C60(0.88) | LDD1656 | [18] |
| LDCM0454 | CL62 | HEK-293T | C60(1.12) | LDD1657 | [18] |
| LDCM0455 | CL63 | HEK-293T | C60(0.85) | LDD1658 | [18] |
| LDCM0456 | CL64 | HEK-293T | C60(0.89) | LDD1659 | [18] |
| LDCM0457 | CL65 | HEK-293T | C60(1.06) | LDD1660 | [18] |
| LDCM0458 | CL66 | HEK-293T | C60(0.78) | LDD1661 | [18] |
| LDCM0459 | CL67 | HEK-293T | C60(1.04) | LDD1662 | [18] |
| LDCM0460 | CL68 | HEK-293T | C60(0.80) | LDD1663 | [18] |
| LDCM0461 | CL69 | HEK-293T | C60(0.94) | LDD1664 | [18] |
| LDCM0462 | CL7 | HEK-293T | C60(1.00) | LDD1665 | [18] |
| LDCM0463 | CL70 | HEK-293T | C60(1.03) | LDD1666 | [18] |
| LDCM0464 | CL71 | HEK-293T | C60(0.95) | LDD1667 | [18] |
| LDCM0465 | CL72 | HEK-293T | C60(1.00) | LDD1668 | [18] |
| LDCM0466 | CL73 | HEK-293T | C60(0.96) | LDD1669 | [18] |
| LDCM0467 | CL74 | HEK-293T | C60(1.11) | LDD1670 | [18] |
| LDCM0469 | CL76 | HEK-293T | C60(1.06) | LDD1672 | [18] |
| LDCM0470 | CL77 | HEK-293T | C60(0.93) | LDD1673 | [18] |
| LDCM0471 | CL78 | HEK-293T | C60(0.91) | LDD1674 | [18] |
| LDCM0472 | CL79 | HEK-293T | C60(1.07) | LDD1675 | [18] |
| LDCM0473 | CL8 | HEK-293T | C60(0.56) | LDD1676 | [18] |
| LDCM0474 | CL80 | HEK-293T | C60(0.81) | LDD1677 | [18] |
| LDCM0475 | CL81 | HEK-293T | C60(0.96) | LDD1678 | [18] |
| LDCM0476 | CL82 | HEK-293T | C60(0.97) | LDD1679 | [18] |
| LDCM0477 | CL83 | HEK-293T | C60(0.93) | LDD1680 | [18] |
| LDCM0478 | CL84 | HEK-293T | C60(0.90) | LDD1681 | [18] |
| LDCM0479 | CL85 | HEK-293T | C60(1.01) | LDD1682 | [18] |
| LDCM0480 | CL86 | HEK-293T | C60(0.92) | LDD1683 | [18] |
| LDCM0481 | CL87 | HEK-293T | C60(0.80) | LDD1684 | [18] |
| LDCM0482 | CL88 | HEK-293T | C60(0.85) | LDD1685 | [18] |
| LDCM0483 | CL89 | HEK-293T | C60(0.96) | LDD1686 | [18] |
| LDCM0484 | CL9 | HEK-293T | C60(0.88) | LDD1687 | [18] |
| LDCM0485 | CL90 | HEK-293T | C60(0.75) | LDD1688 | [18] |
| LDCM0486 | CL91 | HEK-293T | C60(0.84) | LDD1689 | [18] |
| LDCM0487 | CL92 | HEK-293T | C60(0.77) | LDD1690 | [18] |
| LDCM0488 | CL93 | HEK-293T | C60(0.97) | LDD1691 | [18] |
| LDCM0489 | CL94 | HEK-293T | C60(0.96) | LDD1692 | [18] |
| LDCM0490 | CL95 | HEK-293T | C60(0.80) | LDD1693 | [18] |
| LDCM0491 | CL96 | HEK-293T | C60(0.85) | LDD1694 | [18] |
| LDCM0492 | CL97 | HEK-293T | C60(0.88) | LDD1695 | [18] |
| LDCM0493 | CL98 | HEK-293T | C60(0.96) | LDD1696 | [18] |
| LDCM0494 | CL99 | HEK-293T | C60(0.89) | LDD1697 | [18] |
| LDCM0495 | E2913 | HEK-293T | C60(0.96) | LDD1698 | [18] |
| LDCM0625 | F8 | Ramos | C60(0.83) | LDD2187 | [20] |
| LDCM0573 | Fragment11 | Ramos | C60(0.10) | LDD2190 | [20] |
| LDCM0574 | Fragment12 | Ramos | C60(16.33) | LDD2191 | [20] |
| LDCM0575 | Fragment13 | Ramos | C60(0.62) | LDD2192 | [20] |
| LDCM0576 | Fragment14 | Ramos | C60(1.47) | LDD2193 | [20] |
| LDCM0579 | Fragment20 | Ramos | C60(3.00) | LDD2194 | [20] |
| LDCM0580 | Fragment21 | Ramos | C60(7.72) | LDD2195 | [20] |
| LDCM0578 | Fragment27 | Ramos | C60(5.64) | LDD2197 | [20] |
| LDCM0586 | Fragment28 | Ramos | C60(0.54) | LDD2198 | [20] |
| LDCM0588 | Fragment30 | Ramos | C60(9.23) | LDD2199 | [20] |
| LDCM0590 | Fragment32 | Ramos | C60(5.21) | LDD2201 | [20] |
| LDCM0468 | Fragment33 | HEK-293T | C60(0.85) | LDD1671 | [18] |
| LDCM0596 | Fragment38 | Ramos | C60(7.18) | LDD2203 | [20] |
| LDCM0566 | Fragment4 | Ramos | C60(0.86) | LDD2184 | [20] |
| LDCM0427 | Fragment51 | HEK-293T | C60(0.99) | LDD1631 | [18] |
| LDCM0610 | Fragment52 | Ramos | C60(3.02) | LDD2204 | [20] |
| LDCM0614 | Fragment56 | Ramos | C60(2.15) | LDD2205 | [20] |
| LDCM0569 | Fragment7 | Ramos | C60(2.71) | LDD2186 | [20] |
| LDCM0571 | Fragment9 | Ramos | C60(20.00) | LDD2188 | [20] |
| LDCM0116 | HHS-0101 | DM93 | Y245(0.79); Y209(6.96) | LDD0264 | [7] |
| LDCM0117 | HHS-0201 | DM93 | Y245(0.98) | LDD0265 | [7] |
| LDCM0118 | HHS-0301 | DM93 | Y245(2.38); Y209(4.08) | LDD0266 | [7] |
| LDCM0119 | HHS-0401 | DM93 | Y245(1.27); Y209(9.75) | LDD0267 | [7] |
| LDCM0120 | HHS-0701 | DM93 | Y245(0.47); Y209(3.03) | LDD0268 | [7] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [9] |
| LDCM0022 | KB02 | HEK-293T | C60(0.93) | LDD1492 | [18] |
| LDCM0023 | KB03 | HEK-293T | C60(0.96) | LDD1497 | [18] |
| LDCM0024 | KB05 | HMCB | C60(0.84) | LDD3312 | [5] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [9] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Huntingtin (HTT) | Huntingtin family | P42858 | |||
Other
References
