Details of the Target
General Information of Target
| Target ID | LDTP13703 | |||||
|---|---|---|---|---|---|---|
| Target Name | Histone lysine demethylase PHF8 (PHF8) | |||||
| Gene Name | PHF8 | |||||
| Gene ID | 23133 | |||||
| Synonyms |
KIAA1111; ZNF422; Histone lysine demethylase PHF8; EC 1.14.11.27; EC 1.14.11.65; PHD finger protein 8; [histone H3]-dimethyl-L-lysine(36) demethylase PHF8; [histone H3]-dimethyl-L-lysine(9) demethylase PHF8
|
|||||
| 3D Structure | ||||||
| Sequence |
MAENKGGGEAESGGGGSGSAPVTAGAAGPAAQEAEPPLTAVLVEEEEEEGGRAGAEGGAA
GPDDGGVAAASSGSAQAASSPAASVGTGVAGGAVSTPAPAPASAPAPGPSAGPPPGPPAS LLDTCAVCQQSLQSRREAEPKLLPCLHSFCLRCLPEPERQLSVPIPGGSNGDIQQVGVIR CPVCRQECRQIDLVDNYFVKDTSEAPSSSDEKSEQVCTSCEDNASAVGFCVECGEWLCKT CIEAHQRVKFTKDHLIRKKEDVSESVGASGQRPVFCPVHKQEQLKLFCETCDRLTCRDCQ LLEHKEHRYQFLEEAFQNQKGAIENLLAKLLEKKNYVHFAATQVQNRIKEVNETNKRVEQ EIKVAIFTLINEINKKGKSLLQQLENVTKERQMKLLQQQNDITGLSRQVKHVMNFTNWAI ASGSSTALLYSKRLITFQLRHILKARCDPVPAANGAIRFHCDPTFWAKNVVNLGNLVIES KPAPGYTPNVVVGQVPPGTNHISKTPGQINLAQLRLQHMQQQVYAQKHQQLQQMRMQQPP APVPTTTTTTQQHPRQAAPQMLQQQPPRLISVQTMQRGNMNCGAFQAHQMRLAQNAARIP GIPRHSGPQYSMMQPHLQRQHSNPGHAGPFPVVSVHNTTINPTSPTTATMANANRGPTSP SVTAIELIPSVTNPENLPSLPDIPPIQLEDAGSSSLDNLLSRYISGSHLPPQPTSTMNPS PGPSALSPGSSGLSNSHTPVRPPSTSSTGSRGSCGSSGRTAEKTSLSFKSDQVKVKQEPG TEDEICSFSGGVKQEKTEDGRRSACMLSSPESSLTPPLSTNLHLESELDALASLENHVKI EPADMNESCKQSGLSSLVNGKSPIRSLMHRSARIGGDGNNKDDDPNEDWCAVCQNGGDLL CCEKCPKVFHLTCHVPTLLSFPSGDWICTFCRDIGKPEVEYDCDNLQHSKKGKTAQGLSP VDQRKCERLLLYLYCHELSIEFQEPVPASIPNYYKIIKKPMDLSTVKKKLQKKHSQHYQI PDDFVADVRLIFKNCERFNEMMKVVQVYADTQEINLKADSEVAQAGKAVALYFEDKLTEI YSDRTFAPLPEFEQEEDDGEVTEDSDEDFIQPRRKRLKSDERPVHIK |
|||||
| Target Type |
Literature-reported
|
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
JHDM1 histone demethylase family, JHDM1D subfamily
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Histone lysine demethylase with selectivity for the di- and monomethyl states that plays a key role cell cycle progression, rDNA transcription and brain development. Demethylates mono- and dimethylated histone H3 'Lys-9' residue (H3K9Me1 and H3K9Me2), dimethylated H3 'Lys-27' (H3K27Me2) and monomethylated histone H4 'Lys-20' residue (H4K20Me1). Acts as a transcription activator as H3K9Me1, H3K9Me2, H3K27Me2 and H4K20Me1 are epigenetic repressive marks. Involved in cell cycle progression by being required to control G1-S transition. Acts as a coactivator of rDNA transcription, by activating polymerase I (pol I) mediated transcription of rRNA genes. Required for brain development, probably by regulating expression of neuron-specific genes. Only has activity toward H4K20Me1 when nucleosome is used as a substrate and when not histone octamer is used as substrate. May also have weak activity toward dimethylated H3 'Lys-36' (H3K36Me2), however, the relevance of this result remains unsure in vivo. Specifically binds trimethylated 'Lys-4' of histone H3 (H3K4me3), affecting histone demethylase specificity: has weak activity toward H3K9Me2 in absence of H3K4me3, while it has high activity toward H3K9me2 when binding H3K4me3. Positively modulates transcription of histone demethylase KDM5C, acting synergistically with transcription factor ARX; synergy may be related to enrichment of histone H3K4me3 in regulatory elements.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
12.82 | LDD0402 | [1] | |
|
Alkylaryl probe 2 Probe Info |
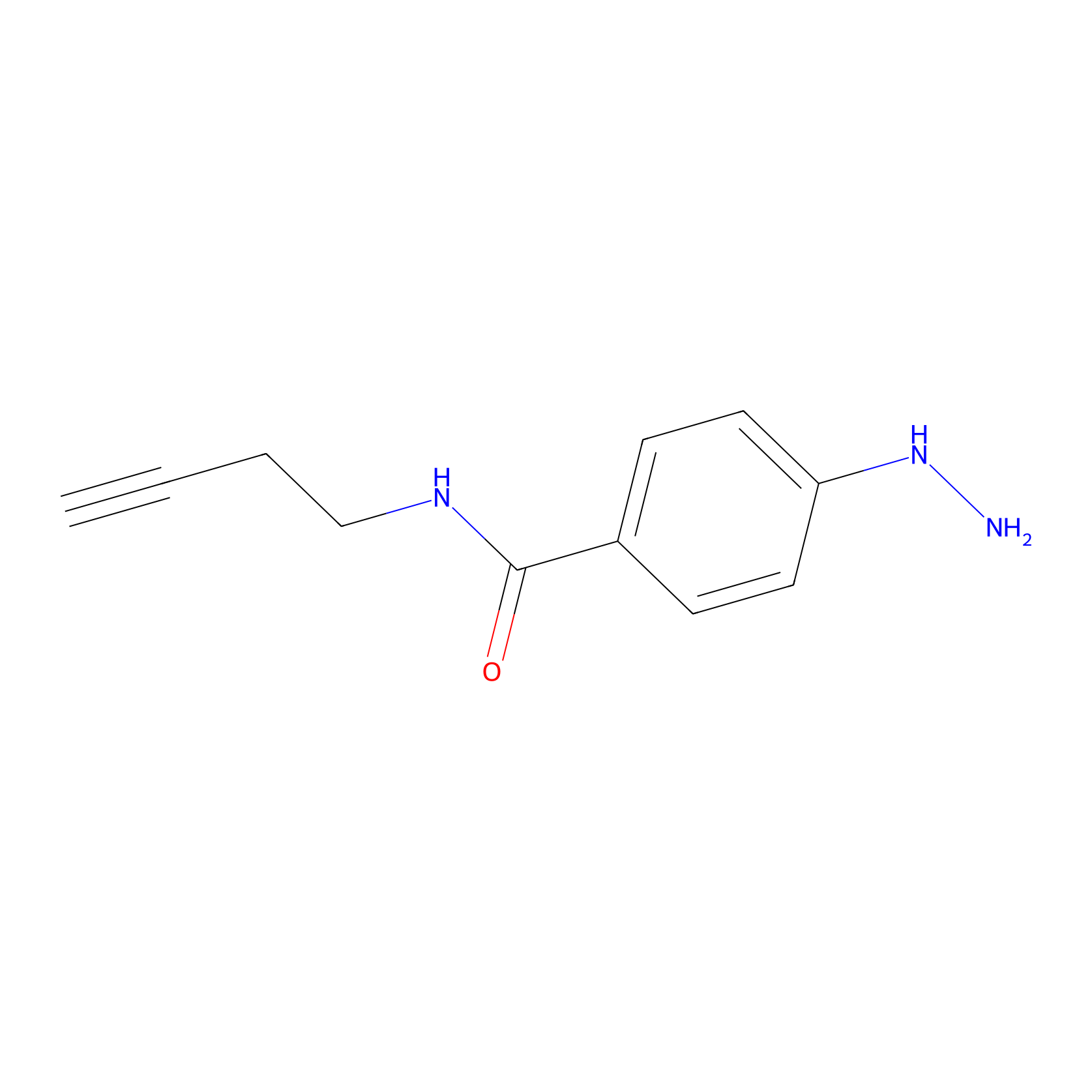 |
20.00 | LDD0389 | [2] | |
|
STPyne Probe Info |
 |
K467(1.19) | LDD0277 | [3] | |
|
DBIA Probe Info |
 |
C271(2.98) | LDD3310 | [4] | |
|
BTD Probe Info |
 |
C684(0.77) | LDD2096 | [5] | |
|
AHL-Pu-1 Probe Info |
 |
C684(2.09) | LDD0169 | [6] | |
|
HHS-475 Probe Info |
 |
Y383(1.22) | LDD0264 | [7] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0221 | [8] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [9] | |
|
IA-alkyne Probe Info |
 |
C684(0.00); C404(0.00) | LDD0036 | [9] | |
|
Lodoacetamide azide Probe Info |
 |
C684(0.00); C260(0.00) | LDD0037 | [9] | |
|
ENE Probe Info |
 |
N.A. | LDD0006 | [10] | |
|
IPM Probe Info |
 |
N.A. | LDD0147 | [11] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [12] | |
|
AOyne Probe Info |
 |
11.70 | LDD0443 | [13] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C170 Probe Info |
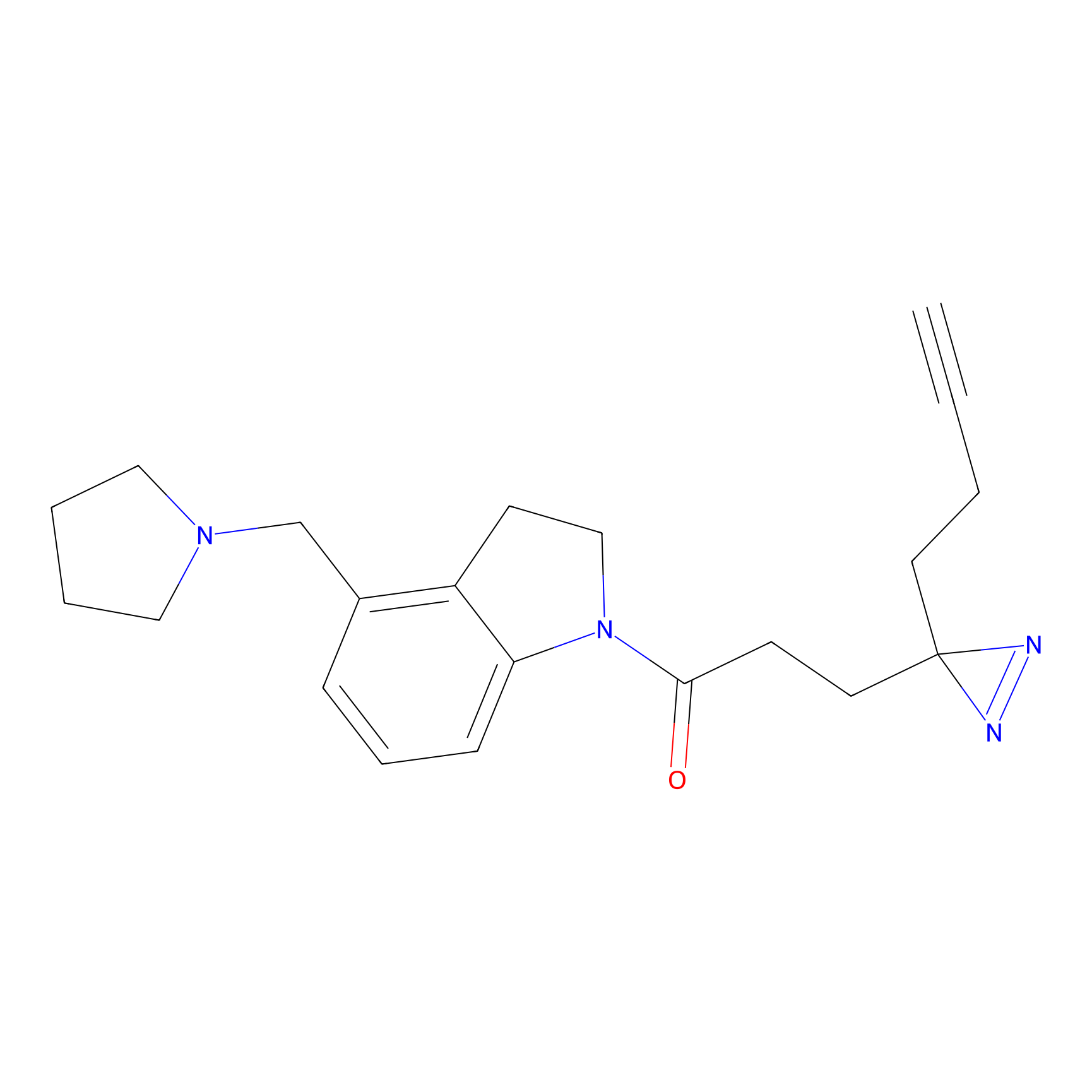 |
10.48 | LDD1850 | [14] | |
|
C361 Probe Info |
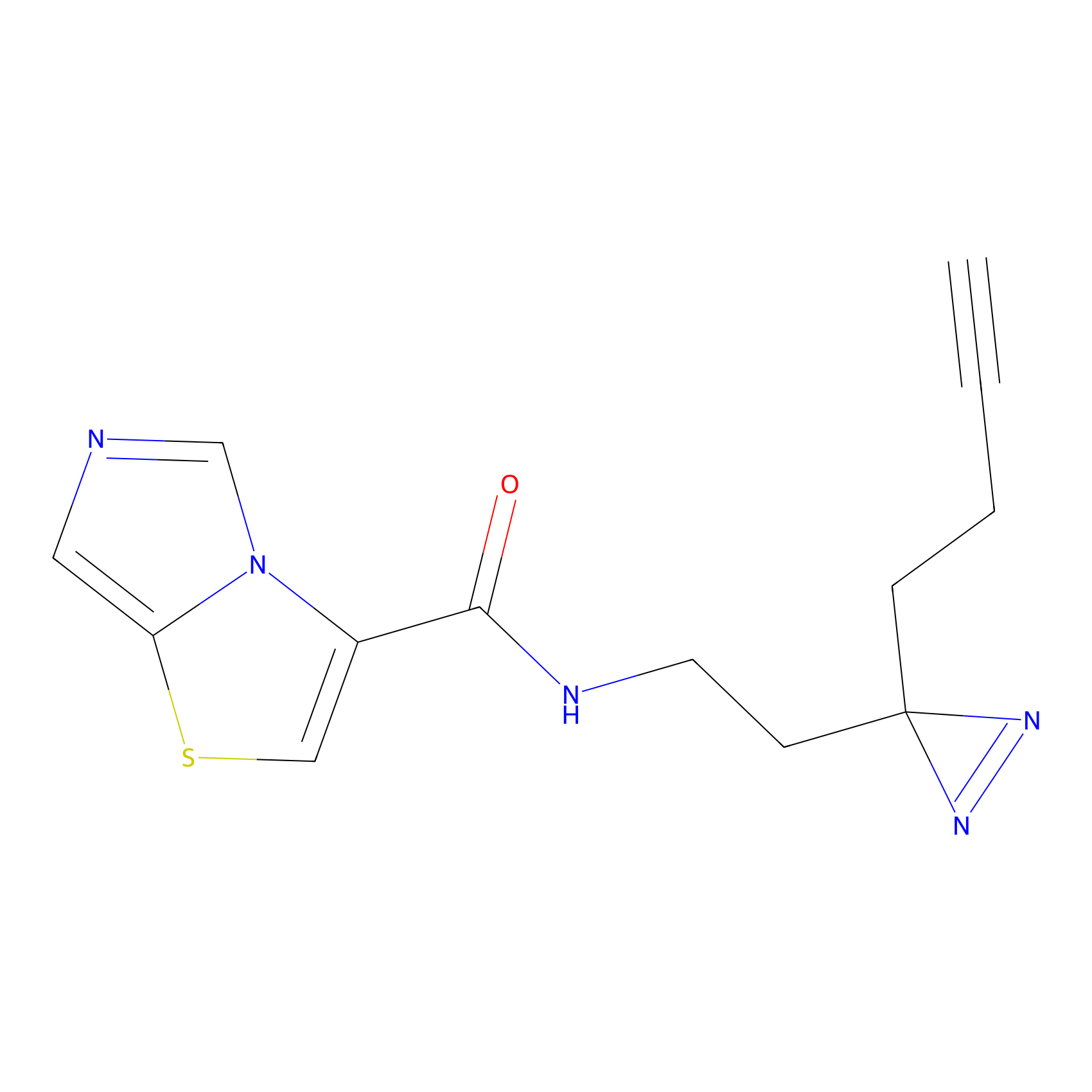 |
20.97 | LDD2022 | [14] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C684(2.09) | LDD0169 | [6] |
| LDCM0214 | AC1 | PaTu 8988t | C404(0.51) | LDD1093 | [15] |
| LDCM0215 | AC10 | PaTu 8988t | C404(0.86) | LDD1094 | [15] |
| LDCM0226 | AC11 | PaTu 8988t | C404(0.70) | LDD1105 | [15] |
| LDCM0237 | AC12 | PaTu 8988t | C404(0.89) | LDD1116 | [15] |
| LDCM0259 | AC14 | PaTu 8988t | C404(1.21) | LDD1138 | [15] |
| LDCM0270 | AC15 | PaTu 8988t | C404(0.79) | LDD1149 | [15] |
| LDCM0276 | AC17 | PaTu 8988t | C260(0.99); C404(0.78) | LDD1155 | [15] |
| LDCM0277 | AC18 | PaTu 8988t | C260(1.01); C404(0.67) | LDD1156 | [15] |
| LDCM0278 | AC19 | PaTu 8988t | C260(0.92); C404(0.68) | LDD1157 | [15] |
| LDCM0279 | AC2 | PaTu 8988t | C404(0.54) | LDD1158 | [15] |
| LDCM0280 | AC20 | PaTu 8988t | C260(0.92); C404(0.57) | LDD1159 | [15] |
| LDCM0281 | AC21 | PaTu 8988t | C260(1.06); C404(0.61) | LDD1160 | [15] |
| LDCM0282 | AC22 | PaTu 8988t | C260(1.06); C404(0.70) | LDD1161 | [15] |
| LDCM0283 | AC23 | PaTu 8988t | C260(0.90); C404(0.70) | LDD1162 | [15] |
| LDCM0284 | AC24 | PaTu 8988t | C260(1.08); C404(0.82) | LDD1163 | [15] |
| LDCM0290 | AC3 | PaTu 8988t | C404(0.60) | LDD1169 | [15] |
| LDCM0296 | AC35 | PaTu 8988t | C260(1.05) | LDD1175 | [15] |
| LDCM0297 | AC36 | PaTu 8988t | C260(1.02) | LDD1176 | [15] |
| LDCM0298 | AC37 | PaTu 8988t | C260(1.03) | LDD1177 | [15] |
| LDCM0299 | AC38 | PaTu 8988t | C260(1.07) | LDD1178 | [15] |
| LDCM0300 | AC39 | PaTu 8988t | C260(0.98) | LDD1179 | [15] |
| LDCM0301 | AC4 | PaTu 8988t | C404(0.65) | LDD1180 | [15] |
| LDCM0302 | AC40 | PaTu 8988t | C260(0.90) | LDD1181 | [15] |
| LDCM0303 | AC41 | PaTu 8988t | C260(1.07) | LDD1182 | [15] |
| LDCM0304 | AC42 | PaTu 8988t | C260(1.06) | LDD1183 | [15] |
| LDCM0305 | AC43 | PaTu 8988t | C260(0.87) | LDD1184 | [15] |
| LDCM0306 | AC44 | PaTu 8988t | C260(1.05) | LDD1185 | [15] |
| LDCM0307 | AC45 | PaTu 8988t | C260(1.02) | LDD1186 | [15] |
| LDCM0308 | AC46 | PaTu 8988t | C260(0.90); C404(0.79) | LDD1187 | [15] |
| LDCM0309 | AC47 | PaTu 8988t | C260(0.80); C404(0.73) | LDD1188 | [15] |
| LDCM0310 | AC48 | PaTu 8988t | C260(0.85); C404(0.59) | LDD1189 | [15] |
| LDCM0311 | AC49 | PaTu 8988t | C260(0.84); C404(0.61) | LDD1190 | [15] |
| LDCM0312 | AC5 | PaTu 8988t | C404(0.53) | LDD1191 | [15] |
| LDCM0313 | AC50 | PaTu 8988t | C260(0.96); C404(0.59) | LDD1192 | [15] |
| LDCM0314 | AC51 | PaTu 8988t | C260(0.31); C404(1.08) | LDD1193 | [15] |
| LDCM0315 | AC52 | PaTu 8988t | C260(0.82); C404(0.64) | LDD1194 | [15] |
| LDCM0316 | AC53 | PaTu 8988t | C260(0.90); C404(0.60) | LDD1195 | [15] |
| LDCM0317 | AC54 | PaTu 8988t | C260(0.98); C404(0.62) | LDD1196 | [15] |
| LDCM0318 | AC55 | PaTu 8988t | C260(0.96); C404(0.77) | LDD1197 | [15] |
| LDCM0319 | AC56 | PaTu 8988t | C260(0.85); C404(1.12) | LDD1198 | [15] |
| LDCM0323 | AC6 | PaTu 8988t | C404(1.26) | LDD1202 | [15] |
| LDCM0332 | AC68 | PaTu 8988t | C404(0.76) | LDD1211 | [15] |
| LDCM0333 | AC69 | PaTu 8988t | C404(0.83) | LDD1212 | [15] |
| LDCM0334 | AC7 | PaTu 8988t | C404(1.07) | LDD1213 | [15] |
| LDCM0335 | AC70 | PaTu 8988t | C404(0.68) | LDD1214 | [15] |
| LDCM0336 | AC71 | PaTu 8988t | C404(0.91) | LDD1215 | [15] |
| LDCM0337 | AC72 | PaTu 8988t | C404(1.07) | LDD1216 | [15] |
| LDCM0338 | AC73 | PaTu 8988t | C404(0.90) | LDD1217 | [15] |
| LDCM0339 | AC74 | PaTu 8988t | C404(0.84) | LDD1218 | [15] |
| LDCM0340 | AC75 | PaTu 8988t | C404(0.56) | LDD1219 | [15] |
| LDCM0341 | AC76 | PaTu 8988t | C404(0.88) | LDD1220 | [15] |
| LDCM0342 | AC77 | PaTu 8988t | C404(0.99) | LDD1221 | [15] |
| LDCM0343 | AC78 | PaTu 8988t | C404(0.67) | LDD1222 | [15] |
| LDCM0344 | AC79 | PaTu 8988t | C404(0.67) | LDD1223 | [15] |
| LDCM0345 | AC8 | PaTu 8988t | C404(1.00) | LDD1224 | [15] |
| LDCM0346 | AC80 | PaTu 8988t | C404(0.83) | LDD1225 | [15] |
| LDCM0347 | AC81 | PaTu 8988t | C404(0.69) | LDD1226 | [15] |
| LDCM0348 | AC82 | PaTu 8988t | C404(0.71) | LDD1227 | [15] |
| LDCM0248 | AKOS034007472 | PaTu 8988t | C404(1.16) | LDD1127 | [15] |
| LDCM0356 | AKOS034007680 | PaTu 8988t | C404(0.84) | LDD1235 | [15] |
| LDCM0275 | AKOS034007705 | PaTu 8988t | C404(0.74) | LDD1154 | [15] |
| LDCM0369 | CL100 | PaTu 8988t | C404(0.59) | LDD1248 | [15] |
| LDCM0370 | CL101 | PaTu 8988t | C404(0.91) | LDD1249 | [15] |
| LDCM0371 | CL102 | PaTu 8988t | C404(0.82) | LDD1250 | [15] |
| LDCM0372 | CL103 | PaTu 8988t | C404(0.86) | LDD1251 | [15] |
| LDCM0373 | CL104 | PaTu 8988t | C404(1.06) | LDD1252 | [15] |
| LDCM0374 | CL105 | PaTu 8988t | C260(0.91); C404(0.77) | LDD1253 | [15] |
| LDCM0375 | CL106 | PaTu 8988t | C260(0.85); C404(1.37) | LDD1254 | [15] |
| LDCM0376 | CL107 | PaTu 8988t | C260(0.91); C404(0.90) | LDD1255 | [15] |
| LDCM0377 | CL108 | PaTu 8988t | C260(0.91); C404(0.76) | LDD1256 | [15] |
| LDCM0378 | CL109 | PaTu 8988t | C260(0.90); C404(0.67) | LDD1257 | [15] |
| LDCM0380 | CL110 | PaTu 8988t | C260(1.02); C404(0.62) | LDD1259 | [15] |
| LDCM0381 | CL111 | PaTu 8988t | C260(1.17); C404(0.64) | LDD1260 | [15] |
| LDCM0387 | CL117 | PaTu 8988t | C260(1.00) | LDD1266 | [15] |
| LDCM0388 | CL118 | PaTu 8988t | C260(1.08) | LDD1267 | [15] |
| LDCM0389 | CL119 | PaTu 8988t | C260(1.04) | LDD1268 | [15] |
| LDCM0391 | CL120 | PaTu 8988t | C260(1.08) | LDD1270 | [15] |
| LDCM0392 | CL121 | PaTu 8988t | C260(0.78); C404(0.77) | LDD1271 | [15] |
| LDCM0393 | CL122 | PaTu 8988t | C260(0.78); C404(2.33) | LDD1272 | [15] |
| LDCM0394 | CL123 | PaTu 8988t | C260(0.82); C404(0.57) | LDD1273 | [15] |
| LDCM0395 | CL124 | PaTu 8988t | C260(0.88); C404(0.55) | LDD1274 | [15] |
| LDCM0453 | CL61 | PaTu 8988t | C260(0.97) | LDD1332 | [15] |
| LDCM0454 | CL62 | PaTu 8988t | C260(1.09) | LDD1333 | [15] |
| LDCM0455 | CL63 | PaTu 8988t | C260(1.03) | LDD1334 | [15] |
| LDCM0456 | CL64 | PaTu 8988t | C260(1.12) | LDD1335 | [15] |
| LDCM0457 | CL65 | PaTu 8988t | C260(0.92) | LDD1336 | [15] |
| LDCM0458 | CL66 | PaTu 8988t | C260(0.83) | LDD1337 | [15] |
| LDCM0459 | CL67 | PaTu 8988t | C260(1.06) | LDD1338 | [15] |
| LDCM0460 | CL68 | PaTu 8988t | C260(1.13) | LDD1339 | [15] |
| LDCM0461 | CL69 | PaTu 8988t | C260(1.01) | LDD1340 | [15] |
| LDCM0463 | CL70 | PaTu 8988t | C260(0.93) | LDD1342 | [15] |
| LDCM0464 | CL71 | PaTu 8988t | C260(0.98) | LDD1343 | [15] |
| LDCM0465 | CL72 | PaTu 8988t | C260(1.39) | LDD1344 | [15] |
| LDCM0466 | CL73 | PaTu 8988t | C260(0.99) | LDD1345 | [15] |
| LDCM0467 | CL74 | PaTu 8988t | C260(1.04) | LDD1346 | [15] |
| LDCM0486 | CL91 | PaTu 8988t | C404(0.76) | LDD1365 | [15] |
| LDCM0487 | CL92 | PaTu 8988t | C404(0.55) | LDD1366 | [15] |
| LDCM0488 | CL93 | PaTu 8988t | C404(0.56) | LDD1367 | [15] |
| LDCM0489 | CL94 | PaTu 8988t | C404(0.84) | LDD1368 | [15] |
| LDCM0490 | CL95 | PaTu 8988t | C404(0.61) | LDD1369 | [15] |
| LDCM0491 | CL96 | PaTu 8988t | C404(0.56) | LDD1370 | [15] |
| LDCM0492 | CL97 | PaTu 8988t | C404(0.63) | LDD1371 | [15] |
| LDCM0493 | CL98 | PaTu 8988t | C404(0.65) | LDD1372 | [15] |
| LDCM0494 | CL99 | PaTu 8988t | C404(0.66) | LDD1373 | [15] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C684(1.44) | LDD1702 | [5] |
| LDCM0468 | Fragment33 | PaTu 8988t | C260(0.93) | LDD1347 | [15] |
| LDCM0116 | HHS-0101 | DM93 | Y383(1.22) | LDD0264 | [7] |
| LDCM0117 | HHS-0201 | DM93 | Y383(6.43) | LDD0265 | [7] |
| LDCM0118 | HHS-0301 | DM93 | Y383(2.91) | LDD0266 | [7] |
| LDCM0119 | HHS-0401 | DM93 | Y383(3.14) | LDD0267 | [7] |
| LDCM0120 | HHS-0701 | DM93 | Y383(3.06) | LDD0268 | [7] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [8] |
| LDCM0022 | KB02 | HEK-293T | C684(1.03); C86(1.00); C260(0.94); C86(1.05) | LDD1492 | [16] |
| LDCM0023 | KB03 | HEK-293T | C684(0.97); C86(0.98); C260(0.92); C86(0.97) | LDD1497 | [16] |
| LDCM0024 | KB05 | COLO792 | C271(2.98) | LDD3310 | [4] |
| LDCM0503 | Nucleophilic fragment 14b | MDA-MB-231 | C684(0.77) | LDD2096 | [5] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C684(1.03) | LDD2111 | [5] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C684(0.95) | LDD2120 | [5] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C684(0.72) | LDD2125 | [5] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C684(0.94) | LDD2128 | [5] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C684(0.94) | LDD2129 | [5] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C684(0.96) | LDD2136 | [5] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C684(0.56) | LDD2140 | [5] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C684(1.35) | LDD2143 | [5] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C684(0.18) | LDD2150 | [5] |
| LDCM0099 | Phenelzine | HEK-293T | 15.00 | LDD0390 | [2] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transcription factor
References
