Details of the Target
General Information of Target
| Target ID | LDTP06631 | |||||
|---|---|---|---|---|---|---|
| Target Name | NADH dehydrogenase 1 alpha subcomplex subunit 9, mitochondrial (NDUFA9) | |||||
| Gene Name | NDUFA9 | |||||
| Gene ID | 4704 | |||||
| Synonyms |
NDUFS2L; NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial; Complex I-39kD; CI-39kD; NADH-ubiquinone oxidoreductase 39 kDa subunit |
|||||
| 3D Structure | ||||||
| Sequence |
MAAAAQSRVVRVLSMSRSAITAIATSVCHGPPCRQLHHALMPHGKGGRSSVSGIVATVFG
ATGFLGRYVVNHLGRMGSQVIIPYRCDKYDIMHLRPMGDLGQLLFLEWDARDKDSIRRVV QHSNVVINLIGRDWETKNFDFEDVFVKIPQAIAQLSKEAGVEKFIHVSHLNANIKSSSRY LRNKAVGEKVVRDAFPEAIIVKPSDIFGREDRFLNSFASMHRFGPIPLGSLGWKTVKQPV YVVDVSKGIVNAVKDPDANGKSFAFVGPSRYLLFHLVKYIFAVAHRLFLPFPLPLFAYRW VARVFEISPFEPWITRDKVERMHITDMKLPHLPGLEDLGIQATPLELKAIEVLRRHRTYR WLSAEIEDVKPAKTVNI |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Complex I NDUFA9 subunit family
|
|||||
| Subcellular location |
Mitochondrion matrix
|
|||||
| Function |
Accessory subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I), that is believed not to be involved in catalysis. Required for proper complex I assembly. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
10.13 | LDD0402 | [1] | |
|
STPyne Probe Info |
 |
K189(1.33); K234(6.82) | LDD0277 | [2] | |
|
IPM Probe Info |
 |
N.A. | LDD0241 | [3] | |
|
BTD Probe Info |
 |
C86(0.88) | LDD2093 | [4] | |
|
Jackson_14 Probe Info |
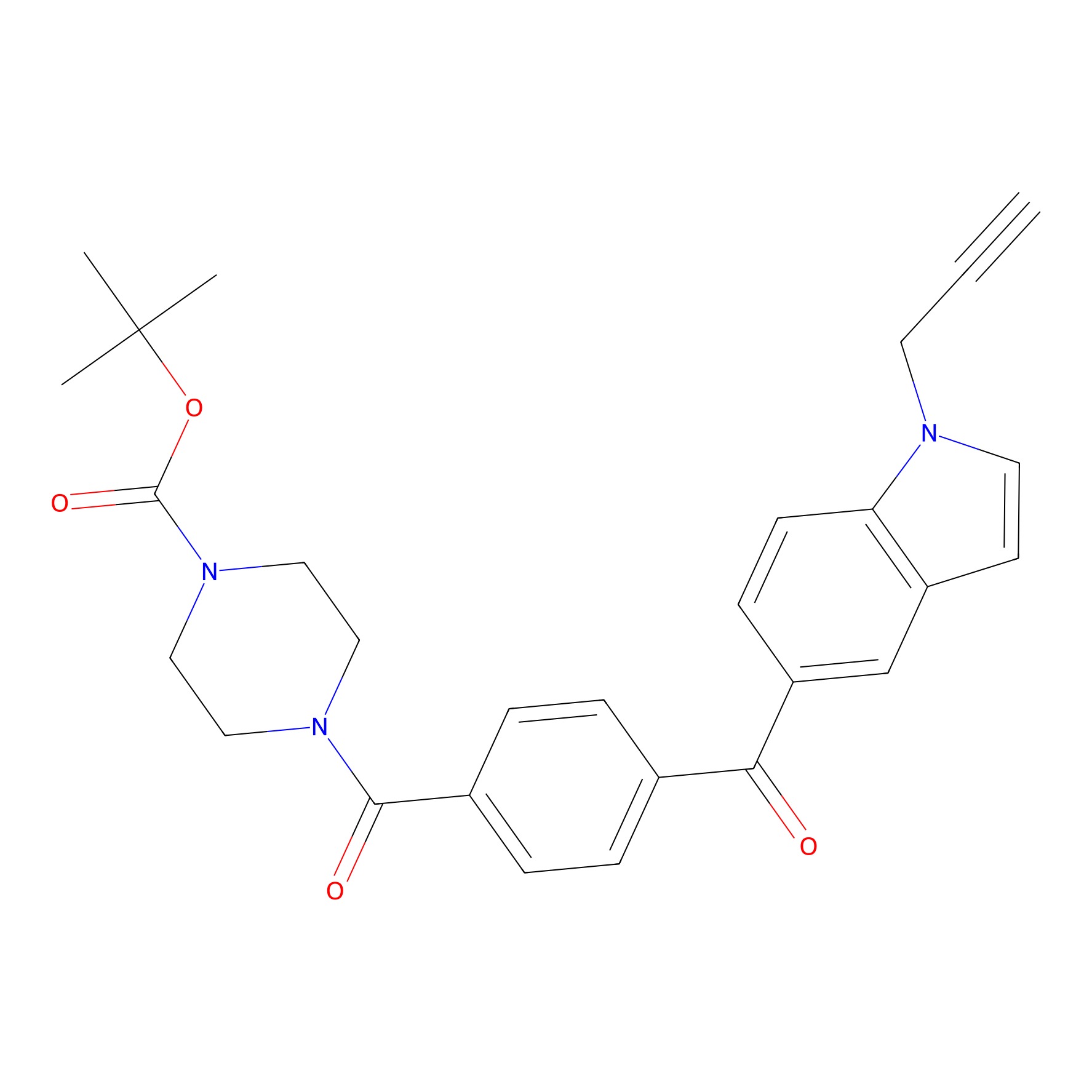 |
2.63 | LDD0123 | [5] | |
|
DBIA Probe Info |
 |
C86(0.93) | LDD1507 | [6] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [7] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [7] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [7] | |
|
Ox-W18 Probe Info |
 |
W313(0.00); W134(0.00) | LDD2175 | [8] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [9] | |
|
AOyne Probe Info |
 |
6.20 | LDD0443 | [10] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe11 Probe Info |
 |
11.68 | LDD0471 | [11] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [11] | |
|
FFF probe14 Probe Info |
 |
20.00 | LDD0477 | [11] | |
|
FFF probe2 Probe Info |
 |
20.00 | LDD0463 | [11] | |
|
FFF probe3 Probe Info |
 |
15.05 | LDD0464 | [11] | |
|
JN0003 Probe Info |
 |
13.71 | LDD0469 | [11] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0136 | [12] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [12] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [13] | |
|
DA-2 Probe Info |
 |
N.A. | LDD0071 | [14] | |
|
OEA-DA Probe Info |
 |
3.13 | LDD0046 | [15] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C86(0.99) | LDD2117 | [4] |
| LDCM0214 | AC1 | HEK-293T | C86(0.93) | LDD1507 | [6] |
| LDCM0259 | AC14 | HEK-293T | C86(1.31) | LDD1512 | [6] |
| LDCM0270 | AC15 | HEK-293T | C86(1.33) | LDD1513 | [6] |
| LDCM0276 | AC17 | HEK-293T | C86(1.23) | LDD1515 | [6] |
| LDCM0281 | AC21 | HEK-293T | C86(1.25) | LDD1520 | [6] |
| LDCM0282 | AC22 | HEK-293T | C86(1.10) | LDD1521 | [6] |
| LDCM0283 | AC23 | HEK-293T | C86(0.95) | LDD1522 | [6] |
| LDCM0284 | AC24 | HEK-293T | C86(0.97) | LDD1523 | [6] |
| LDCM0285 | AC25 | HEK-293T | C86(1.10) | LDD1524 | [6] |
| LDCM0289 | AC29 | HEK-293T | C86(1.24) | LDD1528 | [6] |
| LDCM0291 | AC30 | HEK-293T | C86(1.02) | LDD1530 | [6] |
| LDCM0292 | AC31 | HEK-293T | C86(0.92) | LDD1531 | [6] |
| LDCM0293 | AC32 | HEK-293T | C86(1.77) | LDD1532 | [6] |
| LDCM0294 | AC33 | HEK-293T | C86(1.52) | LDD1533 | [6] |
| LDCM0298 | AC37 | HEK-293T | C86(1.25) | LDD1537 | [6] |
| LDCM0299 | AC38 | HEK-293T | C86(1.49) | LDD1538 | [6] |
| LDCM0300 | AC39 | HEK-293T | C86(1.67) | LDD1539 | [6] |
| LDCM0302 | AC40 | HEK-293T | C86(1.08) | LDD1541 | [6] |
| LDCM0303 | AC41 | HEK-293T | C86(0.88) | LDD1542 | [6] |
| LDCM0307 | AC45 | HEK-293T | C86(1.35) | LDD1546 | [6] |
| LDCM0308 | AC46 | HEK-293T | C86(1.68) | LDD1547 | [6] |
| LDCM0309 | AC47 | HEK-293T | C86(0.94) | LDD1548 | [6] |
| LDCM0310 | AC48 | HEK-293T | C86(1.18) | LDD1549 | [6] |
| LDCM0311 | AC49 | HEK-293T | C86(1.59) | LDD1550 | [6] |
| LDCM0312 | AC5 | HEK-293T | C86(1.28) | LDD1551 | [6] |
| LDCM0316 | AC53 | HEK-293T | C86(1.73) | LDD1555 | [6] |
| LDCM0317 | AC54 | HEK-293T | C86(1.42) | LDD1556 | [6] |
| LDCM0318 | AC55 | HEK-293T | C86(1.21) | LDD1557 | [6] |
| LDCM0319 | AC56 | HEK-293T | C86(1.17) | LDD1558 | [6] |
| LDCM0320 | AC57 | HEK-293T | C86(0.77) | LDD1559 | [6] |
| LDCM0323 | AC6 | HEK-293T | C86(1.27) | LDD1562 | [6] |
| LDCM0325 | AC61 | HEK-293T | C86(0.98) | LDD1564 | [6] |
| LDCM0326 | AC62 | HEK-293T | C86(1.13) | LDD1565 | [6] |
| LDCM0327 | AC63 | HEK-293T | C86(1.05) | LDD1566 | [6] |
| LDCM0328 | AC64 | HEK-293T | C86(0.73) | LDD1567 | [6] |
| LDCM0334 | AC7 | HEK-293T | C86(1.16) | LDD1568 | [6] |
| LDCM0345 | AC8 | HEK-293T | C86(0.84) | LDD1569 | [6] |
| LDCM0248 | AKOS034007472 | HEK-293T | C86(1.13) | LDD1511 | [6] |
| LDCM0356 | AKOS034007680 | HEK-293T | C86(1.23) | LDD1570 | [6] |
| LDCM0275 | AKOS034007705 | HEK-293T | C86(1.18) | LDD1514 | [6] |
| LDCM0156 | Aniline | NCI-H1299 | 11.35 | LDD0403 | [1] |
| LDCM0108 | Chloroacetamide | HeLa | H43(0.00); H122(0.00); H38(0.00) | LDD0222 | [9] |
| LDCM0367 | CL1 | HEK-293T | C86(0.86) | LDD1571 | [6] |
| LDCM0368 | CL10 | HEK-293T | C86(0.44) | LDD1572 | [6] |
| LDCM0370 | CL101 | HEK-293T | C86(1.01) | LDD1574 | [6] |
| LDCM0374 | CL105 | HEK-293T | C86(1.01) | LDD1578 | [6] |
| LDCM0378 | CL109 | HEK-293T | C86(1.08) | LDD1582 | [6] |
| LDCM0379 | CL11 | HEK-293T | C86(0.33) | LDD1583 | [6] |
| LDCM0383 | CL113 | HEK-293T | C86(1.12) | LDD1587 | [6] |
| LDCM0387 | CL117 | HEK-293T | C86(0.95) | LDD1591 | [6] |
| LDCM0390 | CL12 | HEK-293T | C86(0.56) | LDD1594 | [6] |
| LDCM0392 | CL121 | HEK-293T | C86(1.18) | LDD1596 | [6] |
| LDCM0396 | CL125 | HEK-293T | C86(1.10) | LDD1600 | [6] |
| LDCM0400 | CL13 | HEK-293T | C86(0.99) | LDD1604 | [6] |
| LDCM0404 | CL17 | HEK-293T | C86(0.65) | LDD1608 | [6] |
| LDCM0409 | CL21 | HEK-293T | C86(0.53) | LDD1613 | [6] |
| LDCM0410 | CL22 | HEK-293T | C86(0.40) | LDD1614 | [6] |
| LDCM0411 | CL23 | HEK-293T | C86(0.31) | LDD1615 | [6] |
| LDCM0412 | CL24 | HEK-293T | C86(0.37) | LDD1616 | [6] |
| LDCM0413 | CL25 | HEK-293T | C86(0.75) | LDD1617 | [6] |
| LDCM0417 | CL29 | HEK-293T | C86(0.97) | LDD1621 | [6] |
| LDCM0422 | CL33 | HEK-293T | C86(0.51) | LDD1626 | [6] |
| LDCM0423 | CL34 | HEK-293T | C86(0.35) | LDD1627 | [6] |
| LDCM0424 | CL35 | HEK-293T | C86(0.28) | LDD1628 | [6] |
| LDCM0425 | CL36 | HEK-293T | C86(0.54) | LDD1629 | [6] |
| LDCM0426 | CL37 | HEK-293T | C86(0.98) | LDD1630 | [6] |
| LDCM0431 | CL41 | HEK-293T | C86(0.95) | LDD1635 | [6] |
| LDCM0435 | CL45 | HEK-293T | C86(0.61) | LDD1639 | [6] |
| LDCM0436 | CL46 | HEK-293T | C86(0.39) | LDD1640 | [6] |
| LDCM0437 | CL47 | HEK-293T | C86(0.31) | LDD1641 | [6] |
| LDCM0438 | CL48 | HEK-293T | C86(0.31) | LDD1642 | [6] |
| LDCM0439 | CL49 | HEK-293T | C86(1.01) | LDD1643 | [6] |
| LDCM0440 | CL5 | HEK-293T | C86(0.83) | LDD1644 | [6] |
| LDCM0444 | CL53 | HEK-293T | C86(0.81) | LDD1647 | [6] |
| LDCM0448 | CL57 | HEK-293T | C86(0.58) | LDD1651 | [6] |
| LDCM0449 | CL58 | HEK-293T | C86(0.45) | LDD1652 | [6] |
| LDCM0450 | CL59 | HEK-293T | C86(0.45) | LDD1653 | [6] |
| LDCM0452 | CL60 | HEK-293T | C86(0.40) | LDD1655 | [6] |
| LDCM0453 | CL61 | HEK-293T | C86(0.87) | LDD1656 | [6] |
| LDCM0457 | CL65 | HEK-293T | C86(1.17) | LDD1660 | [6] |
| LDCM0461 | CL69 | HEK-293T | C86(0.72) | LDD1664 | [6] |
| LDCM0463 | CL70 | HEK-293T | C86(0.52) | LDD1666 | [6] |
| LDCM0464 | CL71 | HEK-293T | C86(0.51) | LDD1667 | [6] |
| LDCM0465 | CL72 | HEK-293T | C86(0.43) | LDD1668 | [6] |
| LDCM0466 | CL73 | HEK-293T | C86(0.83) | LDD1669 | [6] |
| LDCM0470 | CL77 | HEK-293T | C86(0.40) | LDD1673 | [6] |
| LDCM0475 | CL81 | HEK-293T | C86(0.80) | LDD1678 | [6] |
| LDCM0476 | CL82 | HEK-293T | C86(0.64) | LDD1679 | [6] |
| LDCM0477 | CL83 | HEK-293T | C86(0.51) | LDD1680 | [6] |
| LDCM0478 | CL84 | HEK-293T | C86(0.66) | LDD1681 | [6] |
| LDCM0479 | CL85 | HEK-293T | C86(0.78) | LDD1682 | [6] |
| LDCM0483 | CL89 | HEK-293T | C86(0.68) | LDD1686 | [6] |
| LDCM0484 | CL9 | HEK-293T | C86(0.50) | LDD1687 | [6] |
| LDCM0488 | CL93 | HEK-293T | C86(0.78) | LDD1691 | [6] |
| LDCM0489 | CL94 | HEK-293T | C86(0.64) | LDD1692 | [6] |
| LDCM0490 | CL95 | HEK-293T | C86(0.67) | LDD1693 | [6] |
| LDCM0491 | CL96 | HEK-293T | C86(0.90) | LDD1694 | [6] |
| LDCM0492 | CL97 | HEK-293T | C86(0.82) | LDD1695 | [6] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [9] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C86(0.82) | LDD2121 | [4] |
| LDCM0109 | NEM | HeLa | H122(0.00); H38(0.00) | LDD0223 | [9] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C86(0.88) | LDD2093 | [4] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C86(0.86) | LDD2099 | [4] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C86(1.10) | LDD2107 | [4] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C86(0.94) | LDD2108 | [4] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C86(0.67) | LDD2109 | [4] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C86(1.01) | LDD2111 | [4] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C86(2.10) | LDD2119 | [4] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C86(0.80) | LDD2125 | [4] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C86(1.21) | LDD2129 | [4] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C86(1.21) | LDD2136 | [4] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C86(1.90) | LDD2144 | [4] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C86(0.89) | LDD2146 | [4] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C86(1.01) | LDD2153 | [4] |
| LDCM0016 | Ranjitkar_cp1 | MDA-MB-231 | 2.63 | LDD0123 | [5] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Nitric oxide-associated protein 1 (NOA1) | TRAFAC class YlqF/YawG GTPase family | Q8NC60 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Huntingtin (HTT) | Huntingtin family | P42858 | |||
Other
References
