Details of the Target
General Information of Target
| Target ID | LDTP11764 | |||||
|---|---|---|---|---|---|---|
| Target Name | Nuclear ubiquitous casein and cyclin-dependent kinase substrate 1 (NUCKS1) | |||||
| Gene Name | NUCKS1 | |||||
| Gene ID | 64710 | |||||
| Synonyms |
NUCKS; Nuclear ubiquitous casein and cyclin-dependent kinase substrate 1; P1 |
|||||
| 3D Structure | ||||||
| Sequence |
MAPARAGFCPLLLLLLLGLWVAEIPVSAKPKGMTSSQWFKIQHMQPSPQACNSAMKNINK
HTKRCKDLNTFLHEPFSSVAATCQTPKIACKNGDKNCHQSHGAVSLTMCKLTSGKHPNCR YKEKRQNKSYVVACKPPQKKDSQQFHLVPVHLDRVL |
|||||
| Target Bioclass |
Other
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Chromatin-associated protein involved in DNA repair by promoting homologous recombination (HR). Binds double-stranded DNA (dsDNA) and secondary DNA structures, such as D-loop structures, but with less affinity than RAD51AP1.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C-Sul Probe Info |
 |
11.81 | LDD0066 | [1] | |
|
ONAyne Probe Info |
 |
K87(10.00) | LDD0274 | [2] | |
|
STPyne Probe Info |
 |
K171(9.19); K175(7.99); K184(2.90); K188(6.67) | LDD0277 | [2] | |
|
AZ-9 Probe Info |
 |
E18(1.00) | LDD2208 | [3] | |
|
Probe 1 Probe Info |
 |
Y13(13.05); Y26(57.38) | LDD3495 | [4] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0221 | [5] | |
|
AMP probe Probe Info |
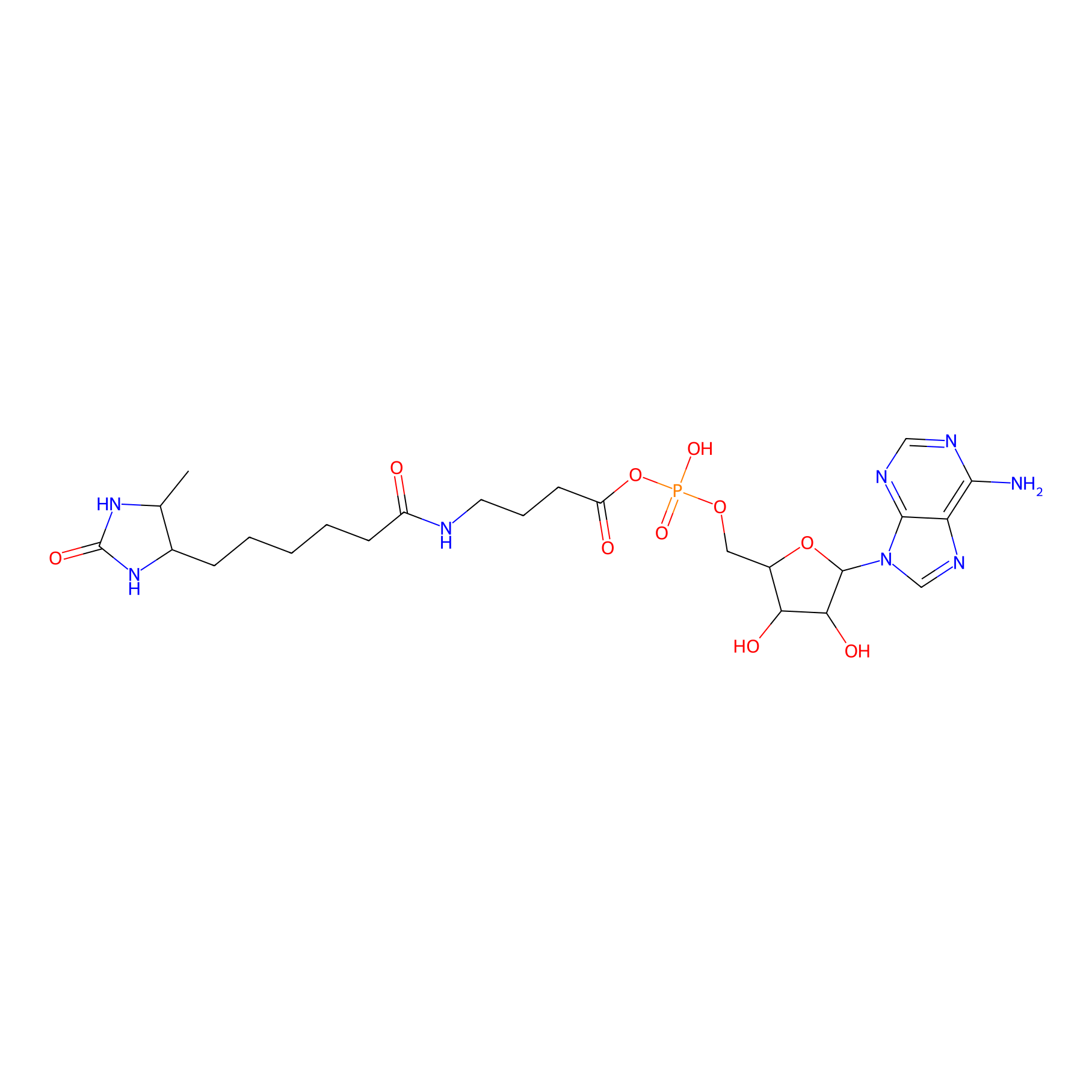 |
K175(0.00); K184(0.00); K186(0.00); K9(0.00) | LDD0200 | [6] | |
|
ATP probe Probe Info |
 |
K175(0.00); K9(0.00); K184(0.00); K186(0.00) | LDD0199 | [6] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [7] | |
|
NHS Probe Info |
 |
K184(0.00); K175(0.00); K9(0.00) | LDD0010 | [8] | |
|
SF Probe Info |
 |
Y26(0.00); K159(0.00); Y13(0.00) | LDD0028 | [9] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [10] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0357 | [10] | |
|
HHS-465 Probe Info |
 |
K9(0.00); K98(0.00); Y13(0.00); K188(0.00) | LDD2240 | [11] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C187 Probe Info |
 |
19.03 | LDD1865 | [12] | |
|
C191 Probe Info |
 |
11.16 | LDD1868 | [12] | |
|
C193 Probe Info |
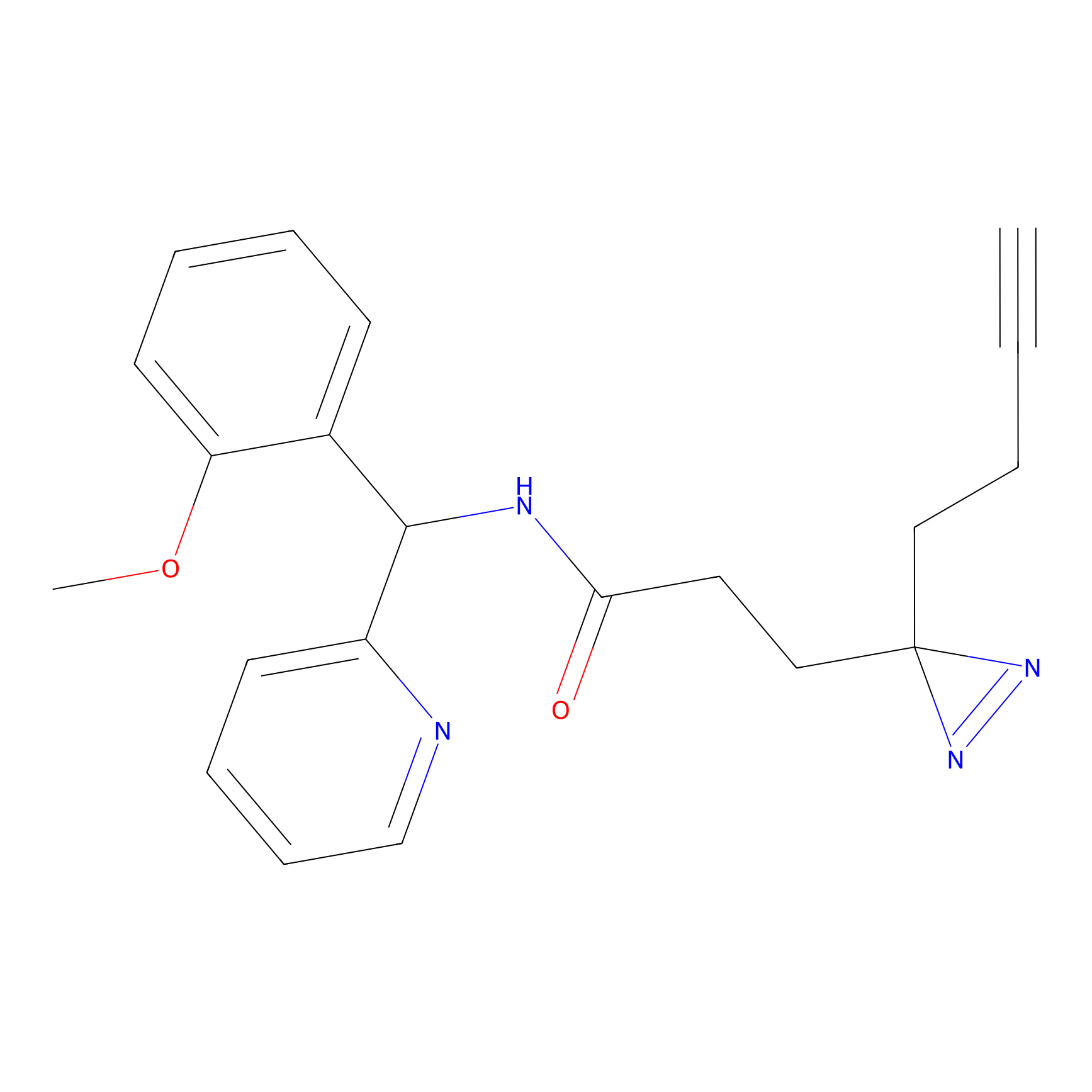 |
5.10 | LDD1869 | [12] | |
|
C354 Probe Info |
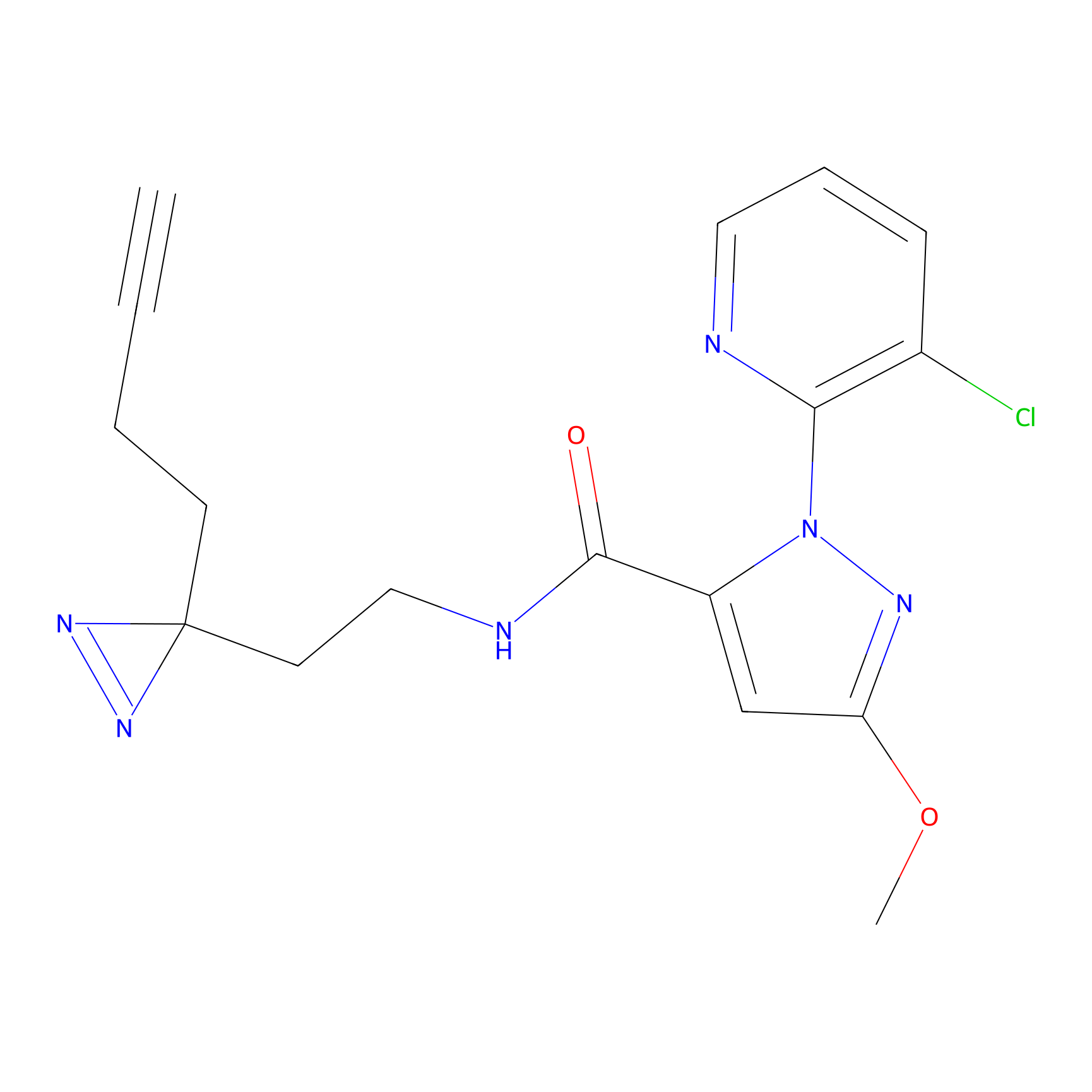 |
8.57 | LDD2015 | [12] | |
Competitor(s) Related to This Target
References
