Details of the Target
General Information of Target
| Target ID | LDTP11117 | |||||
|---|---|---|---|---|---|---|
| Target Name | Serine/arginine-rich splicing factor 8 (SRSF8) | |||||
| Gene Name | SRSF8 | |||||
| Gene ID | 10929 | |||||
| Synonyms |
SFRS2B; SRP46; Serine/arginine-rich splicing factor 8; Pre-mRNA-splicing factor SRP46; Splicing factor SRp46; Splicing factor, arginine/serine-rich 2B |
|||||
| 3D Structure | ||||||
| Sequence |
MVWPWVAMASRWGPLIGLAPCCLWLLGAVLLMDASARPANHSSTRERVANREENEILPPD
HLNGVKLEMDGHLNRGFHQEVFLGKDLGGFDEDAEPRRSRRKLMVIFSKVDVNTDRKISA KEMQRWIMEKTAEHFQEAMEESKTHFRAVDPDGDGHVSWDEYKVKFLASKGHSEKEVADA IRLNEELKVDEETQEVLENLKDRWYQADSPPADLLLTEEEFLSFLHPEHSRGMLRFMVKE IVRDLDQDGDKQLSVPEFISLPVGTVENQQGQDIDDNWVKDRKKEFEELIDSNHDGIVTA EELESYMDPMNEYNALNEAKQMIAVADENQNHHLEPEEVLKYSEFFTGSKLVDYARSVHE EF |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Splicing factor SR family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function | Involved in pre-mRNA alternative splicing. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| AGS | SNV: p.F62L | . | |||
| AN3CA | Deletion: p.P7LfsTer7 | . | |||
| HEC1 | Substitution: p.P128Y | DBIA Probe Info | |||
| HEC1B | Substitution: p.P128Y | . | |||
| RPMI8226 | SNV: p.S210L | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DBIA Probe Info |
 |
C141(1.29) | LDD3332 | [1] | |
|
HHS-482 Probe Info |
 |
Y44(0.96) | LDD0285 | [2] | |
|
HHS-475 Probe Info |
 |
Y23(0.38); Y44(1.05) | LDD0264 | [3] | |
|
HHS-465 Probe Info |
 |
Y23(7.71); Y44(6.08) | LDD2237 | [4] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0165 | [5] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2224 | [6] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [7] | |
|
OSF Probe Info |
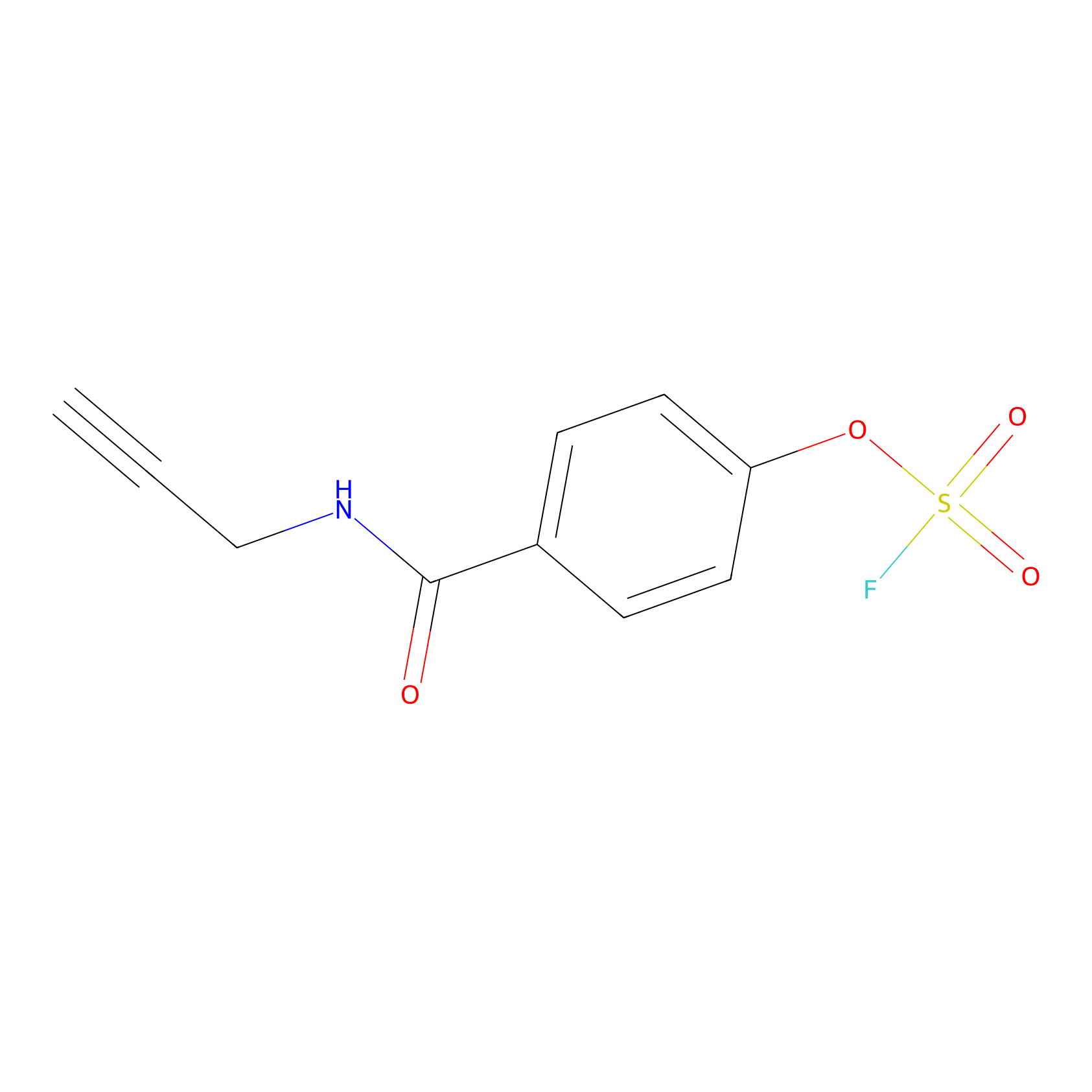 |
N.A. | LDD0029 | [8] | |
|
SF Probe Info |
 |
Y185(0.00); Y44(0.00); Y23(0.00) | LDD0028 | [8] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [7] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [9] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0116 | HHS-0101 | DM93 | Y23(0.38); Y44(1.05) | LDD0264 | [3] |
| LDCM0117 | HHS-0201 | DM93 | Y23(0.51); Y44(0.91) | LDD0265 | [3] |
| LDCM0118 | HHS-0301 | DM93 | Y23(0.50); Y44(0.95) | LDD0266 | [3] |
| LDCM0119 | HHS-0401 | DM93 | Y23(0.41); Y44(1.08) | LDD0267 | [3] |
| LDCM0120 | HHS-0701 | DM93 | Y23(0.33); Y44(0.91) | LDD0268 | [3] |
| LDCM0123 | JWB131 | DM93 | Y44(0.96) | LDD0285 | [2] |
| LDCM0124 | JWB142 | DM93 | Y44(0.65) | LDD0286 | [2] |
| LDCM0125 | JWB146 | DM93 | Y44(1.21) | LDD0287 | [2] |
| LDCM0126 | JWB150 | DM93 | Y44(2.63) | LDD0288 | [2] |
| LDCM0127 | JWB152 | DM93 | Y44(1.60) | LDD0289 | [2] |
| LDCM0128 | JWB198 | DM93 | Y44(0.94) | LDD0290 | [2] |
| LDCM0129 | JWB202 | DM93 | Y44(0.36) | LDD0291 | [2] |
| LDCM0130 | JWB211 | DM93 | Y44(0.91) | LDD0292 | [2] |
| LDCM0022 | KB02 | CCK-81 | C141(2.32) | LDD2297 | [1] |
| LDCM0023 | KB03 | CCK-81 | C141(1.82) | LDD2714 | [1] |
| LDCM0024 | KB05 | MKN45 | C141(1.29) | LDD3332 | [1] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Other
References
