Details of the Target
General Information of Target
| Target ID | LDTP11046 | |||||
|---|---|---|---|---|---|---|
| Target Name | RNA-binding protein 4B (RBM4B) | |||||
| Gene Name | RBM4B | |||||
| Gene ID | 83759 | |||||
| Synonyms |
RBM30; RNA-binding protein 4B; RNA-binding motif protein 30; RNA-binding motif protein 4B; RNA-binding protein 30 |
|||||
| 3D Structure | ||||||
| Sequence |
MAFLDNPTIILAHIRQSHVTSDDTGMCEMVLIDHDVDLEKIHPPSMPGDSGSEIQGSNGE
TQGYVYAQSVDITSSWDFGIRRRSNTAQRLERLRKERQNQIKCKNIQWKERNSKQSAQEL KSLFEKKSLKEKPPISGKQSILSVRLEQCPLQLNNPFNEYSKFDGKGHVGTTATKKIDVY LPLHSSQDRLLPMTVVTMASARVQDLIGLICWQYTSEGREPKLNDNVSAYCLHIAEDDGE VDTDFPPLDSNEPIHKFGFSTLALVEKYSSPGLTSKESLFVRINAAHGFSLIQVDNTKVT MKEILLKAVKRRKGSQKVSGPQYRLEKQSEPNVAVDLDSTLESQSAWEFCLVRENSSRAD GVFEEDSQIDIATVQDMLSSHHYKSFKVSMIHRLRFTTDVQLGISGDKVEIDPVTNQKAS TKFWIKQKPISIDSDLLCACDLAEEKSPSHAIFKLTYLSNHDYKHLYFESDAATVNEIVL KVNYILESRASTARADYFAQKQRKLNRRTSFSFQKEKKSGQQ |
|||||
| Target Bioclass |
Other
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function | Required for the translational activation of PER1 mRNA in response to circadian clock. Binds directly to the 3'-UTR of the PER1 mRNA. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K3(8.33) | LDD0277 | [1] | |
|
IPM Probe Info |
 |
C31(0.00); C162(0.00); C107(0.00); C89(0.00) | LDD0241 | [2] | |
|
Probe 1 Probe Info |
 |
Y25(43.87); Y101(16.16); Y194(13.00); Y320(8.66) | LDD3495 | [3] | |
|
DBIA Probe Info |
 |
C107(1.56); C31(1.34) | LDD3312 | [4] | |
|
BTD Probe Info |
 |
C31(0.65) | LDD2090 | [5] | |
|
AHL-Pu-1 Probe Info |
 |
C107(2.36) | LDD0168 | [6] | |
|
HPAP Probe Info |
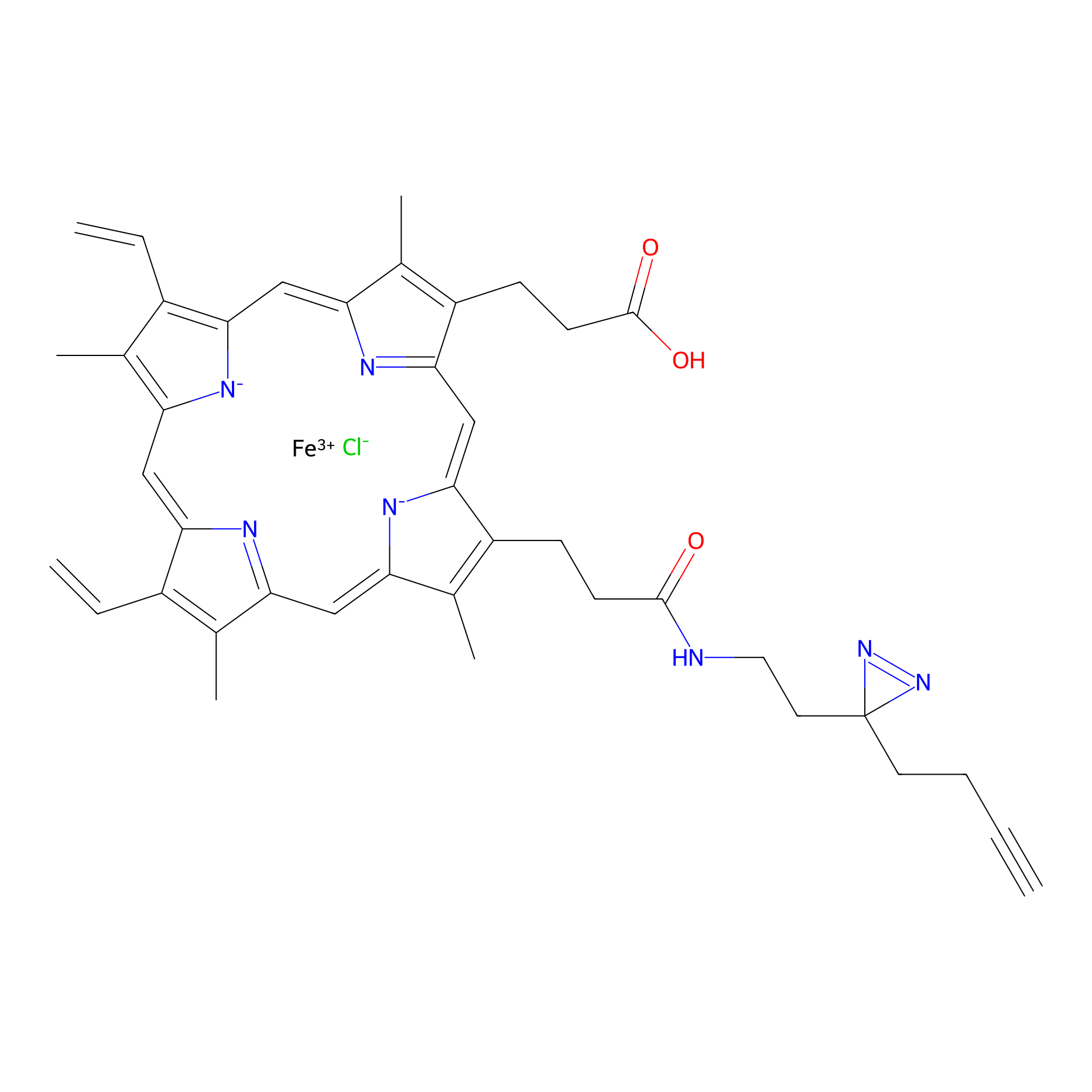 |
3.75 | LDD0062 | [7] | |
|
HHS-465 Probe Info |
 |
Y37(6.87); Y58(10.00) | LDD2237 | [8] | |
|
ATP probe Probe Info |
 |
K71(0.00); K3(0.00); K35(0.00); K79(0.00) | LDD0199 | [9] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [10] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [10] | |
|
WYneN Probe Info |
 |
C31(0.00); C162(0.00) | LDD0021 | [11] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [11] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [11] | |
|
Phosphinate-6 Probe Info |
 |
C175(0.00); C162(0.00); C165(0.00); C31(0.00) | LDD0018 | [12] | |
|
Acrolein Probe Info |
 |
H41(0.00); C31(0.00); C89(0.00); C107(0.00) | LDD0217 | [13] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [13] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [13] | |
|
NAIA_5 Probe Info |
 |
C107(0.00); C165(0.00); C89(0.00); C162(0.00) | LDD2223 | [14] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe11 Probe Info |
 |
8.16 | LDD0471 | [15] | |
|
FFF probe13 Probe Info |
 |
16.95 | LDD0475 | [15] | |
|
FFF probe2 Probe Info |
 |
16.98 | LDD0463 | [15] | |
|
FFF probe3 Probe Info |
 |
13.29 | LDD0464 | [15] | |
|
FFF probe6 Probe Info |
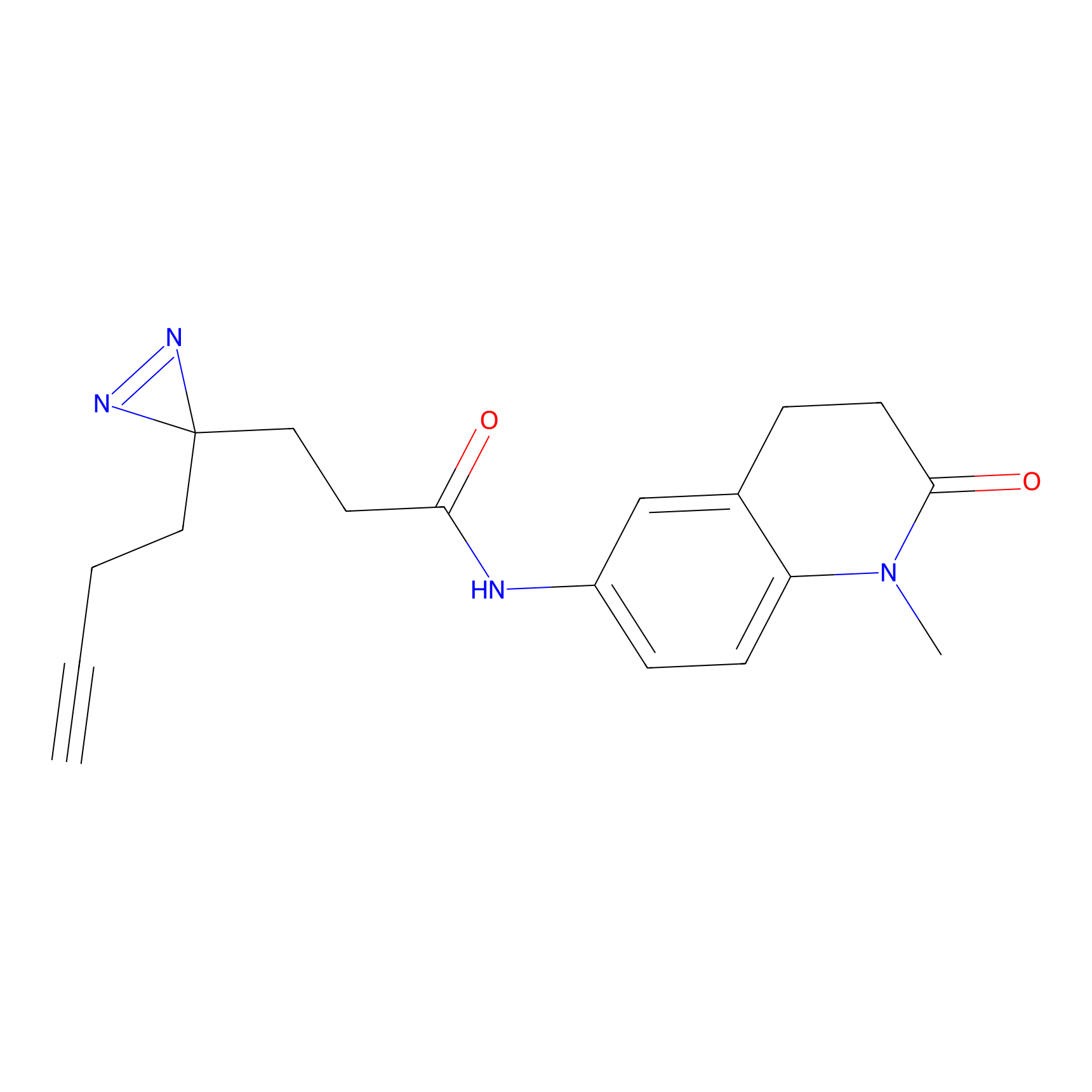 |
20.00 | LDD0467 | [15] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C31(0.65) | LDD2112 | [5] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C31(1.04); C89(0.81) | LDD2117 | [5] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C31(0.96) | LDD2103 | [5] |
| LDCM0025 | 4SU-RNA | HEK-293T | C107(2.36) | LDD0168 | [6] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C107(2.73) | LDD0169 | [6] |
| LDCM0214 | AC1 | HEK-293T | C89(0.75) | LDD1507 | [16] |
| LDCM0276 | AC17 | HEK-293T | C89(1.07) | LDD1515 | [16] |
| LDCM0284 | AC24 | HEK-293T | C89(0.89) | LDD1523 | [16] |
| LDCM0285 | AC25 | HEK-293T | C89(0.90) | LDD1524 | [16] |
| LDCM0293 | AC32 | HEK-293T | C89(0.89) | LDD1532 | [16] |
| LDCM0294 | AC33 | HEK-293T | C89(0.82) | LDD1533 | [16] |
| LDCM0302 | AC40 | HEK-293T | C89(1.07) | LDD1541 | [16] |
| LDCM0303 | AC41 | HEK-293T | C89(1.06) | LDD1542 | [16] |
| LDCM0310 | AC48 | HEK-293T | C89(1.05) | LDD1549 | [16] |
| LDCM0311 | AC49 | HEK-293T | C89(1.04) | LDD1550 | [16] |
| LDCM0319 | AC56 | HEK-293T | C89(0.99) | LDD1558 | [16] |
| LDCM0320 | AC57 | HEK-293T | C89(1.30) | LDD1559 | [16] |
| LDCM0328 | AC64 | HEK-293T | C89(1.09) | LDD1567 | [16] |
| LDCM0345 | AC8 | HEK-293T | C89(0.87) | LDD1569 | [16] |
| LDCM0356 | AKOS034007680 | HEK-293T | C89(1.10) | LDD1570 | [16] |
| LDCM0275 | AKOS034007705 | HEK-293T | C89(0.99) | LDD1514 | [16] |
| LDCM0108 | Chloroacetamide | HeLa | H41(0.00); C162(0.00); C107(0.00) | LDD0222 | [13] |
| LDCM0367 | CL1 | HEK-293T | C89(1.13) | LDD1571 | [16] |
| LDCM0370 | CL101 | HEK-293T | C89(1.05) | LDD1574 | [16] |
| LDCM0374 | CL105 | HEK-293T | C89(0.97) | LDD1578 | [16] |
| LDCM0378 | CL109 | HEK-293T | C89(1.06) | LDD1582 | [16] |
| LDCM0383 | CL113 | HEK-293T | C89(1.12) | LDD1587 | [16] |
| LDCM0387 | CL117 | HEK-293T | C89(1.13) | LDD1591 | [16] |
| LDCM0390 | CL12 | HEK-293T | C89(0.84) | LDD1594 | [16] |
| LDCM0392 | CL121 | HEK-293T | C89(0.91) | LDD1596 | [16] |
| LDCM0396 | CL125 | HEK-293T | C89(1.00) | LDD1600 | [16] |
| LDCM0400 | CL13 | HEK-293T | C89(0.96) | LDD1604 | [16] |
| LDCM0404 | CL17 | HEK-293T | C89(0.96) | LDD1608 | [16] |
| LDCM0412 | CL24 | HEK-293T | C89(1.03) | LDD1616 | [16] |
| LDCM0413 | CL25 | HEK-293T | C89(0.88) | LDD1617 | [16] |
| LDCM0417 | CL29 | HEK-293T | C89(0.89) | LDD1621 | [16] |
| LDCM0425 | CL36 | HEK-293T | C89(0.99) | LDD1629 | [16] |
| LDCM0426 | CL37 | HEK-293T | C89(1.10) | LDD1630 | [16] |
| LDCM0431 | CL41 | HEK-293T | C89(0.93) | LDD1635 | [16] |
| LDCM0438 | CL48 | HEK-293T | C89(0.99) | LDD1642 | [16] |
| LDCM0439 | CL49 | HEK-293T | C89(1.05) | LDD1643 | [16] |
| LDCM0440 | CL5 | HEK-293T | C89(0.89) | LDD1644 | [16] |
| LDCM0444 | CL53 | HEK-293T | C89(0.82) | LDD1647 | [16] |
| LDCM0452 | CL60 | HEK-293T | C89(1.27) | LDD1655 | [16] |
| LDCM0453 | CL61 | HEK-293T | C89(1.01) | LDD1656 | [16] |
| LDCM0457 | CL65 | HEK-293T | C89(1.01) | LDD1660 | [16] |
| LDCM0465 | CL72 | HEK-293T | C89(1.00) | LDD1668 | [16] |
| LDCM0466 | CL73 | HEK-293T | C89(0.96) | LDD1669 | [16] |
| LDCM0470 | CL77 | HEK-293T | C89(0.80) | LDD1673 | [16] |
| LDCM0478 | CL84 | HEK-293T | C89(0.90) | LDD1681 | [16] |
| LDCM0479 | CL85 | HEK-293T | C89(1.09) | LDD1682 | [16] |
| LDCM0483 | CL89 | HEK-293T | C89(0.76) | LDD1686 | [16] |
| LDCM0491 | CL96 | HEK-293T | C89(0.90) | LDD1694 | [16] |
| LDCM0492 | CL97 | HEK-293T | C89(0.89) | LDD1695 | [16] |
| LDCM0625 | F8 | Ramos | C89(0.86) | LDD2187 | [17] |
| LDCM0576 | Fragment14 | Ramos | C89(1.71) | LDD2193 | [17] |
| LDCM0586 | Fragment28 | Ramos | C89(0.59) | LDD2198 | [17] |
| LDCM0566 | Fragment4 | Ramos | C89(0.53) | LDD2184 | [17] |
| LDCM0569 | Fragment7 | Ramos | C89(0.45) | LDD2186 | [17] |
| LDCM0107 | IAA | HeLa | C107(0.00); H41(0.00); H117(0.00); C89(0.00) | LDD0221 | [13] |
| LDCM0022 | KB02 | HEK-293T | C89(1.10) | LDD1492 | [16] |
| LDCM0023 | KB03 | HEK-293T | C89(1.02) | LDD1497 | [16] |
| LDCM0024 | KB05 | HMCB | C107(1.56); C31(1.34) | LDD3312 | [4] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C31(0.91) | LDD2102 | [5] |
| LDCM0109 | NEM | HeLa | H41(0.00); H61(0.00); H142(0.00) | LDD0223 | [13] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C31(0.65) | LDD2090 | [5] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C31(0.44) | LDD2106 | [5] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C31(1.01) | LDD2114 | [5] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C31(0.48) | LDD2141 | [5] |
| LDCM0551 | Nucleophilic fragment 5b | MDA-MB-231 | C31(2.34) | LDD2145 | [5] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C31(0.87) | LDD2153 | [5] |
| LDCM0014 | Panhematin | HEK-293T | 3.75 | LDD0062 | [7] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Histone-lysine N-methyltransferase SMYD1 (SMYD1) | Class V-like SAM-binding methyltransferase superfamily | Q8NB12 | |||
| E3 ubiquitin-protein ligase DZIP3 (DZIP3) | . | Q86Y13 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Krueppel-like factor 1 (KLF1) | Krueppel C2H2-type zinc-finger protein family | Q13351 | |||
| NF-kappa-B inhibitor delta (NFKBID) | NF-kappa-B inhibitor family | Q8NI38 | |||
Other
References
