Details of the Target
General Information of Target
| Target ID | LDTP08957 | |||||
|---|---|---|---|---|---|---|
| Target Name | Tumor suppressor ARF (CDKN2A) | |||||
| Gene Name | CDKN2A | |||||
| Gene ID | 1029 | |||||
| Synonyms |
CDKN2; MLM; Tumor suppressor ARF; Alternative reading frame; ARF; Cyclin-dependent kinase inhibitor 2A; p14ARF |
|||||
| 3D Structure | ||||||
| Sequence |
MVRRFLVTLRIRRACGPPRVRVFVVHIPRLTGEWAAPGAPAAVALVLMLLRSQRLGQQPL
PRRPGHDDGQRPSGGAAAAPRRGAQLRRPRHSHPTRARRCPGGLPGHAGGAAPGRGAAGR ARCLGPSARGPG |
|||||
| Target Bioclass |
Other
|
|||||
| Subcellular location |
Nucleus, nucleolus; Mitochondrion
|
|||||
| Function |
Capable of inducing cell cycle arrest in G1 and G2 phases. Acts as a tumor suppressor. Binds to MDM2 and blocks its nucleocytoplasmic shuttling by sequestering it in the nucleolus. This inhibits the oncogenic action of MDM2 by blocking MDM2-induced degradation of p53 and enhancing p53-dependent transactivation and apoptosis. Also induces G2 arrest and apoptosis in a p53-independent manner by preventing the activation of cyclin B1/CDC2 complexes. Binds to BCL6 and down-regulates BCL6-induced transcriptional repression. Binds to E2F1 and MYC and blocks their transcriptional activator activity but has no effect on MYC transcriptional repression. Binds to TOP1/TOPOI and stimulates its activity. This complex binds to rRNA gene promoters and may play a role in rRNA transcription and/or maturation. Interacts with NPM1/B23 and promotes its polyubiquitination and degradation, thus inhibiting rRNA processing. Plays a role in inhibiting ribosome biogenesis, perhaps by binding to the nucleolar localization sequence of transcription termination factor TTF1, and thereby preventing nucleolar localization of TTF1. Interacts with COMMD1 and promotes its 'Lys63'-linked polyubiquitination. Interacts with UBE2I/UBC9 and enhances sumoylation of a number of its binding partners including MDM2 and E2F1. Binds to HUWE1 and represses its ubiquitin ligase activity. May play a role in controlling cell proliferation and apoptosis during mammary gland development.; [Isoform smARF]: May be involved in regulation of autophagy and caspase-independent cell death; the short-lived mitochondrial isoform is stabilized by C1QBP.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
AHL-Pu-1 Probe Info |
 |
C100(2.03) | LDD0169 | [2] | |
|
DBIA Probe Info |
 |
C100(0.95) | LDD0078 | [3] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0165 | [4] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [5] | |
|
TFBX Probe Info |
 |
C100(0.00); C123(0.00) | LDD0027 | [5] | |
|
IPM Probe Info |
 |
C123(0.00); C100(0.00) | LDD0005 | [6] | |
|
NPM Probe Info |
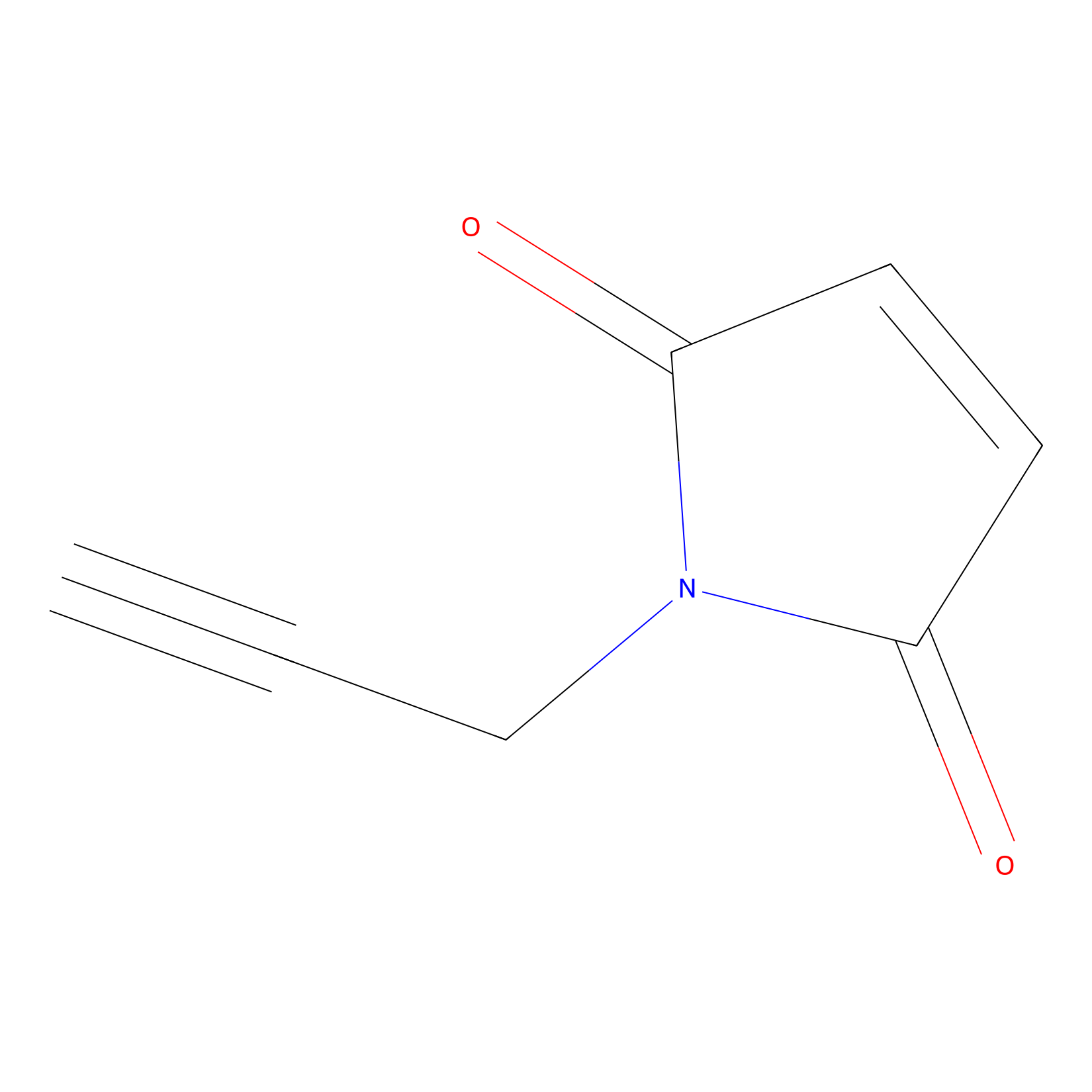 |
N.A. | LDD0016 | [6] | |
|
Phosphinate-6 Probe Info |
 |
C100(0.00); C123(0.00) | LDD0018 | [7] | |
|
Acrolein Probe Info |
 |
H66(0.00); C100(0.00) | LDD0217 | [8] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [8] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [8] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [9] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C100(2.03) | LDD0169 | [2] |
| LDCM0214 | AC1 | HEK-293T | C100(0.91); C123(0.99) | LDD1507 | [10] |
| LDCM0215 | AC10 | PaTu 8988t | C100(1.15) | LDD1094 | [3] |
| LDCM0226 | AC11 | PaTu 8988t | C100(1.36) | LDD1105 | [3] |
| LDCM0237 | AC12 | PaTu 8988t | C100(1.16) | LDD1116 | [3] |
| LDCM0246 | AC128 | PaTu 8988t | C100(1.13) | LDD1125 | [3] |
| LDCM0247 | AC129 | PaTu 8988t | C100(1.01) | LDD1126 | [3] |
| LDCM0249 | AC130 | PaTu 8988t | C100(1.10) | LDD1128 | [3] |
| LDCM0250 | AC131 | PaTu 8988t | C100(1.10) | LDD1129 | [3] |
| LDCM0251 | AC132 | PaTu 8988t | C100(1.28) | LDD1130 | [3] |
| LDCM0252 | AC133 | PaTu 8988t | C100(1.30) | LDD1131 | [3] |
| LDCM0253 | AC134 | PaTu 8988t | C100(0.99) | LDD1132 | [3] |
| LDCM0254 | AC135 | PaTu 8988t | C100(1.60) | LDD1133 | [3] |
| LDCM0255 | AC136 | PaTu 8988t | C100(1.19) | LDD1134 | [3] |
| LDCM0256 | AC137 | PaTu 8988t | C100(1.31) | LDD1135 | [3] |
| LDCM0257 | AC138 | PaTu 8988t | C100(0.97) | LDD1136 | [3] |
| LDCM0258 | AC139 | PaTu 8988t | C100(0.88) | LDD1137 | [3] |
| LDCM0259 | AC14 | PaTu 8988t | C100(0.94) | LDD1138 | [3] |
| LDCM0260 | AC140 | PaTu 8988t | C100(1.09) | LDD1139 | [3] |
| LDCM0261 | AC141 | PaTu 8988t | C100(1.04) | LDD1140 | [3] |
| LDCM0262 | AC142 | PaTu 8988t | C100(0.91) | LDD1141 | [3] |
| LDCM0270 | AC15 | PaTu 8988t | C100(1.07) | LDD1149 | [3] |
| LDCM0276 | AC17 | PaTu 8988t | C100(0.90) | LDD1155 | [3] |
| LDCM0277 | AC18 | PaTu 8988t | C100(1.10) | LDD1156 | [3] |
| LDCM0278 | AC19 | PaTu 8988t | C100(0.89) | LDD1157 | [3] |
| LDCM0279 | AC2 | HEK-293T | C100(0.89); C123(0.92) | LDD1518 | [10] |
| LDCM0280 | AC20 | PaTu 8988t | C100(1.43) | LDD1159 | [3] |
| LDCM0281 | AC21 | PaTu 8988t | C100(1.18) | LDD1160 | [3] |
| LDCM0282 | AC22 | PaTu 8988t | C100(1.45) | LDD1161 | [3] |
| LDCM0283 | AC23 | PaTu 8988t | C100(1.69) | LDD1162 | [3] |
| LDCM0284 | AC24 | PaTu 8988t | C100(1.30) | LDD1163 | [3] |
| LDCM0285 | AC25 | PaTu 8988t | C100(1.60) | LDD1164 | [3] |
| LDCM0286 | AC26 | PaTu 8988t | C100(1.39) | LDD1165 | [3] |
| LDCM0287 | AC27 | PaTu 8988t | C100(1.70) | LDD1166 | [3] |
| LDCM0288 | AC28 | PaTu 8988t | C100(1.94) | LDD1167 | [3] |
| LDCM0289 | AC29 | PaTu 8988t | C100(1.44) | LDD1168 | [3] |
| LDCM0290 | AC3 | HEK-293T | C100(0.92); C123(0.98) | LDD1529 | [10] |
| LDCM0291 | AC30 | PaTu 8988t | C100(1.50) | LDD1170 | [3] |
| LDCM0292 | AC31 | PaTu 8988t | C100(1.57) | LDD1171 | [3] |
| LDCM0293 | AC32 | PaTu 8988t | C100(1.38) | LDD1172 | [3] |
| LDCM0294 | AC33 | PaTu 8988t | C100(1.71) | LDD1173 | [3] |
| LDCM0295 | AC34 | PaTu 8988t | C100(1.22) | LDD1174 | [3] |
| LDCM0296 | AC35 | PaTu 8988t | C100(1.40) | LDD1175 | [3] |
| LDCM0297 | AC36 | PaTu 8988t | C100(1.67) | LDD1176 | [3] |
| LDCM0298 | AC37 | PaTu 8988t | C100(1.42) | LDD1177 | [3] |
| LDCM0299 | AC38 | PaTu 8988t | C100(1.27) | LDD1178 | [3] |
| LDCM0300 | AC39 | PaTu 8988t | C100(1.50) | LDD1179 | [3] |
| LDCM0301 | AC4 | HEK-293T | C100(1.01); C123(0.98) | LDD1540 | [10] |
| LDCM0302 | AC40 | PaTu 8988t | C100(1.54) | LDD1181 | [3] |
| LDCM0303 | AC41 | PaTu 8988t | C100(1.24) | LDD1182 | [3] |
| LDCM0304 | AC42 | PaTu 8988t | C100(1.16) | LDD1183 | [3] |
| LDCM0305 | AC43 | PaTu 8988t | C100(1.27) | LDD1184 | [3] |
| LDCM0306 | AC44 | PaTu 8988t | C100(1.48) | LDD1185 | [3] |
| LDCM0307 | AC45 | PaTu 8988t | C100(1.15) | LDD1186 | [3] |
| LDCM0308 | AC46 | PaTu 8988t | C100(1.14) | LDD1187 | [3] |
| LDCM0309 | AC47 | PaTu 8988t | C100(1.30) | LDD1188 | [3] |
| LDCM0310 | AC48 | PaTu 8988t | C100(1.37) | LDD1189 | [3] |
| LDCM0311 | AC49 | PaTu 8988t | C100(1.59) | LDD1190 | [3] |
| LDCM0312 | AC5 | HEK-293T | C100(1.00); C123(0.99) | LDD1551 | [10] |
| LDCM0313 | AC50 | PaTu 8988t | C100(1.68) | LDD1192 | [3] |
| LDCM0314 | AC51 | PaTu 8988t | C100(0.78) | LDD1193 | [3] |
| LDCM0315 | AC52 | PaTu 8988t | C100(1.27) | LDD1194 | [3] |
| LDCM0316 | AC53 | PaTu 8988t | C100(1.90) | LDD1195 | [3] |
| LDCM0317 | AC54 | PaTu 8988t | C100(2.01) | LDD1196 | [3] |
| LDCM0318 | AC55 | PaTu 8988t | C100(1.20) | LDD1197 | [3] |
| LDCM0319 | AC56 | PaTu 8988t | C100(1.68) | LDD1198 | [3] |
| LDCM0320 | AC57 | PaTu 8988t | C100(1.36) | LDD1199 | [3] |
| LDCM0321 | AC58 | PaTu 8988t | C100(1.32) | LDD1200 | [3] |
| LDCM0322 | AC59 | PaTu 8988t | C100(1.38) | LDD1201 | [3] |
| LDCM0323 | AC6 | PaTu 8988t | C100(1.13) | LDD1202 | [3] |
| LDCM0324 | AC60 | PaTu 8988t | C100(1.32) | LDD1203 | [3] |
| LDCM0325 | AC61 | PaTu 8988t | C100(0.87) | LDD1204 | [3] |
| LDCM0326 | AC62 | PaTu 8988t | C100(0.98) | LDD1205 | [3] |
| LDCM0327 | AC63 | PaTu 8988t | C100(0.84) | LDD1206 | [3] |
| LDCM0328 | AC64 | PaTu 8988t | C100(0.89) | LDD1207 | [3] |
| LDCM0329 | AC65 | PaTu 8988t | C100(0.72) | LDD1208 | [3] |
| LDCM0330 | AC66 | PaTu 8988t | C100(0.83) | LDD1209 | [3] |
| LDCM0331 | AC67 | PaTu 8988t | C100(1.00) | LDD1210 | [3] |
| LDCM0332 | AC68 | PaTu 8988t | C100(0.98) | LDD1211 | [3] |
| LDCM0333 | AC69 | PaTu 8988t | C100(1.02) | LDD1212 | [3] |
| LDCM0334 | AC7 | PaTu 8988t | C100(0.97) | LDD1213 | [3] |
| LDCM0335 | AC70 | PaTu 8988t | C100(1.14) | LDD1214 | [3] |
| LDCM0336 | AC71 | PaTu 8988t | C100(1.24) | LDD1215 | [3] |
| LDCM0337 | AC72 | PaTu 8988t | C100(1.06) | LDD1216 | [3] |
| LDCM0338 | AC73 | PaTu 8988t | C100(0.98) | LDD1217 | [3] |
| LDCM0339 | AC74 | PaTu 8988t | C100(1.30) | LDD1218 | [3] |
| LDCM0340 | AC75 | PaTu 8988t | C100(1.49) | LDD1219 | [3] |
| LDCM0341 | AC76 | PaTu 8988t | C100(1.10) | LDD1220 | [3] |
| LDCM0342 | AC77 | PaTu 8988t | C100(1.20) | LDD1221 | [3] |
| LDCM0343 | AC78 | PaTu 8988t | C100(1.25) | LDD1222 | [3] |
| LDCM0344 | AC79 | PaTu 8988t | C100(1.55) | LDD1223 | [3] |
| LDCM0345 | AC8 | PaTu 8988t | C100(1.02) | LDD1224 | [3] |
| LDCM0346 | AC80 | PaTu 8988t | C100(1.22) | LDD1225 | [3] |
| LDCM0347 | AC81 | PaTu 8988t | C100(1.60) | LDD1226 | [3] |
| LDCM0348 | AC82 | PaTu 8988t | C100(1.49) | LDD1227 | [3] |
| LDCM0349 | AC83 | PaTu 8988t | C100(0.92) | LDD1228 | [3] |
| LDCM0350 | AC84 | PaTu 8988t | C100(0.83) | LDD1229 | [3] |
| LDCM0351 | AC85 | PaTu 8988t | C100(1.06) | LDD1230 | [3] |
| LDCM0352 | AC86 | PaTu 8988t | C100(1.00) | LDD1231 | [3] |
| LDCM0353 | AC87 | PaTu 8988t | C100(0.96) | LDD1232 | [3] |
| LDCM0354 | AC88 | PaTu 8988t | C100(0.95) | LDD1233 | [3] |
| LDCM0355 | AC89 | PaTu 8988t | C100(0.84) | LDD1234 | [3] |
| LDCM0357 | AC90 | PaTu 8988t | C100(0.95) | LDD1236 | [3] |
| LDCM0358 | AC91 | PaTu 8988t | C100(0.92) | LDD1237 | [3] |
| LDCM0359 | AC92 | PaTu 8988t | C100(0.92) | LDD1238 | [3] |
| LDCM0360 | AC93 | PaTu 8988t | C100(0.84) | LDD1239 | [3] |
| LDCM0361 | AC94 | PaTu 8988t | C100(0.87) | LDD1240 | [3] |
| LDCM0362 | AC95 | PaTu 8988t | C100(0.85) | LDD1241 | [3] |
| LDCM0363 | AC96 | PaTu 8988t | C100(0.77) | LDD1242 | [3] |
| LDCM0364 | AC97 | PaTu 8988t | C100(0.74) | LDD1243 | [3] |
| LDCM0248 | AKOS034007472 | PaTu 8988t | C100(1.06) | LDD1127 | [3] |
| LDCM0356 | AKOS034007680 | PaTu 8988t | C100(1.47) | LDD1235 | [3] |
| LDCM0275 | AKOS034007705 | PaTu 8988t | C100(1.11) | LDD1154 | [3] |
| LDCM0156 | Aniline | NCI-H1299 | 11.68 | LDD0403 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C100(0.95) | LDD0078 | [3] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [8] |
| LDCM0367 | CL1 | HEK-293T | C100(0.89); C123(1.04); C15(0.90) | LDD1571 | [10] |
| LDCM0368 | CL10 | HEK-293T | C100(1.13); C123(0.96) | LDD1572 | [10] |
| LDCM0369 | CL100 | HEK-293T | C100(1.05); C123(0.92) | LDD1573 | [10] |
| LDCM0370 | CL101 | PaTu 8988t | C100(1.21) | LDD1249 | [3] |
| LDCM0371 | CL102 | PaTu 8988t | C100(1.19) | LDD1250 | [3] |
| LDCM0372 | CL103 | PaTu 8988t | C100(1.34) | LDD1251 | [3] |
| LDCM0373 | CL104 | PaTu 8988t | C100(1.18) | LDD1252 | [3] |
| LDCM0374 | CL105 | PaTu 8988t | C100(1.05) | LDD1253 | [3] |
| LDCM0375 | CL106 | PaTu 8988t | C100(0.96) | LDD1254 | [3] |
| LDCM0376 | CL107 | PaTu 8988t | C100(1.09) | LDD1255 | [3] |
| LDCM0377 | CL108 | PaTu 8988t | C100(1.27) | LDD1256 | [3] |
| LDCM0378 | CL109 | PaTu 8988t | C100(1.26) | LDD1257 | [3] |
| LDCM0379 | CL11 | HEK-293T | C100(1.09); C123(0.92) | LDD1583 | [10] |
| LDCM0380 | CL110 | PaTu 8988t | C100(1.60) | LDD1259 | [3] |
| LDCM0381 | CL111 | PaTu 8988t | C100(1.66) | LDD1260 | [3] |
| LDCM0382 | CL112 | PaTu 8988t | C100(1.35) | LDD1261 | [3] |
| LDCM0383 | CL113 | PaTu 8988t | C100(1.24) | LDD1262 | [3] |
| LDCM0384 | CL114 | PaTu 8988t | C100(0.96) | LDD1263 | [3] |
| LDCM0385 | CL115 | PaTu 8988t | C100(1.09) | LDD1264 | [3] |
| LDCM0386 | CL116 | PaTu 8988t | C100(1.50) | LDD1265 | [3] |
| LDCM0387 | CL117 | PaTu 8988t | C100(1.06) | LDD1266 | [3] |
| LDCM0388 | CL118 | PaTu 8988t | C100(1.07) | LDD1267 | [3] |
| LDCM0389 | CL119 | PaTu 8988t | C100(1.21) | LDD1268 | [3] |
| LDCM0390 | CL12 | HEK-293T | C100(1.18); C123(0.94) | LDD1594 | [10] |
| LDCM0391 | CL120 | PaTu 8988t | C100(1.39) | LDD1270 | [3] |
| LDCM0392 | CL121 | PaTu 8988t | C100(1.31) | LDD1271 | [3] |
| LDCM0393 | CL122 | PaTu 8988t | C100(1.76) | LDD1272 | [3] |
| LDCM0394 | CL123 | PaTu 8988t | C100(1.58) | LDD1273 | [3] |
| LDCM0395 | CL124 | PaTu 8988t | C100(1.18) | LDD1274 | [3] |
| LDCM0396 | CL125 | PaTu 8988t | C100(0.97) | LDD1275 | [3] |
| LDCM0397 | CL126 | PaTu 8988t | C100(1.06) | LDD1276 | [3] |
| LDCM0398 | CL127 | PaTu 8988t | C100(1.21) | LDD1277 | [3] |
| LDCM0399 | CL128 | PaTu 8988t | C100(1.23) | LDD1278 | [3] |
| LDCM0400 | CL13 | HEK-293T | C100(1.01); C123(1.20); C15(0.77) | LDD1604 | [10] |
| LDCM0401 | CL14 | HEK-293T | C100(1.18); C123(0.93) | LDD1605 | [10] |
| LDCM0402 | CL15 | HEK-293T | C100(1.18); C123(1.00) | LDD1606 | [10] |
| LDCM0403 | CL16 | PaTu 8988t | C100(1.16) | LDD1282 | [3] |
| LDCM0404 | CL17 | PaTu 8988t | C100(1.15) | LDD1283 | [3] |
| LDCM0405 | CL18 | PaTu 8988t | C100(1.18) | LDD1284 | [3] |
| LDCM0406 | CL19 | PaTu 8988t | C100(1.00) | LDD1285 | [3] |
| LDCM0407 | CL2 | HEK-293T | C100(1.08); C123(0.94) | LDD1611 | [10] |
| LDCM0408 | CL20 | PaTu 8988t | C100(1.23) | LDD1287 | [3] |
| LDCM0409 | CL21 | PaTu 8988t | C100(1.30) | LDD1288 | [3] |
| LDCM0410 | CL22 | PaTu 8988t | C100(1.25) | LDD1289 | [3] |
| LDCM0411 | CL23 | PaTu 8988t | C100(1.15) | LDD1290 | [3] |
| LDCM0412 | CL24 | PaTu 8988t | C100(0.96) | LDD1291 | [3] |
| LDCM0413 | CL25 | PaTu 8988t | C100(0.96) | LDD1292 | [3] |
| LDCM0414 | CL26 | PaTu 8988t | C100(1.13) | LDD1293 | [3] |
| LDCM0415 | CL27 | PaTu 8988t | C100(0.98) | LDD1294 | [3] |
| LDCM0416 | CL28 | PaTu 8988t | C100(1.24) | LDD1295 | [3] |
| LDCM0417 | CL29 | PaTu 8988t | C100(1.16) | LDD1296 | [3] |
| LDCM0418 | CL3 | HEK-293T | C100(1.01); C123(0.88) | LDD1622 | [10] |
| LDCM0419 | CL30 | PaTu 8988t | C100(1.17) | LDD1298 | [3] |
| LDCM0420 | CL31 | PaTu 8988t | C100(1.08) | LDD1299 | [3] |
| LDCM0421 | CL32 | PaTu 8988t | C100(1.51) | LDD1300 | [3] |
| LDCM0422 | CL33 | PaTu 8988t | C100(1.71) | LDD1301 | [3] |
| LDCM0423 | CL34 | PaTu 8988t | C100(1.62) | LDD1302 | [3] |
| LDCM0424 | CL35 | PaTu 8988t | C100(1.43) | LDD1303 | [3] |
| LDCM0425 | CL36 | PaTu 8988t | C100(1.50) | LDD1304 | [3] |
| LDCM0426 | CL37 | PaTu 8988t | C100(1.83) | LDD1305 | [3] |
| LDCM0428 | CL39 | PaTu 8988t | C100(1.72) | LDD1307 | [3] |
| LDCM0429 | CL4 | HEK-293T | C100(0.98); C123(0.89) | LDD1633 | [10] |
| LDCM0430 | CL40 | PaTu 8988t | C100(1.75) | LDD1309 | [3] |
| LDCM0431 | CL41 | PaTu 8988t | C100(1.82) | LDD1310 | [3] |
| LDCM0432 | CL42 | PaTu 8988t | C100(2.18) | LDD1311 | [3] |
| LDCM0433 | CL43 | PaTu 8988t | C100(2.51) | LDD1312 | [3] |
| LDCM0434 | CL44 | PaTu 8988t | C100(1.82) | LDD1313 | [3] |
| LDCM0435 | CL45 | PaTu 8988t | C100(1.55) | LDD1314 | [3] |
| LDCM0436 | CL46 | PaTu 8988t | C100(1.10) | LDD1315 | [3] |
| LDCM0437 | CL47 | PaTu 8988t | C100(1.30) | LDD1316 | [3] |
| LDCM0438 | CL48 | PaTu 8988t | C100(1.29) | LDD1317 | [3] |
| LDCM0439 | CL49 | PaTu 8988t | C100(1.19) | LDD1318 | [3] |
| LDCM0440 | CL5 | HEK-293T | C100(0.92); C123(1.01) | LDD1644 | [10] |
| LDCM0441 | CL50 | PaTu 8988t | C100(1.17) | LDD1320 | [3] |
| LDCM0442 | CL51 | PaTu 8988t | C100(1.16) | LDD1321 | [3] |
| LDCM0443 | CL52 | PaTu 8988t | C100(1.16) | LDD1322 | [3] |
| LDCM0444 | CL53 | PaTu 8988t | C100(1.44) | LDD1323 | [3] |
| LDCM0445 | CL54 | PaTu 8988t | C100(1.45) | LDD1324 | [3] |
| LDCM0446 | CL55 | PaTu 8988t | C100(1.41) | LDD1325 | [3] |
| LDCM0447 | CL56 | PaTu 8988t | C100(0.72) | LDD1326 | [3] |
| LDCM0448 | CL57 | PaTu 8988t | C100(1.54) | LDD1327 | [3] |
| LDCM0449 | CL58 | PaTu 8988t | C100(1.36) | LDD1328 | [3] |
| LDCM0450 | CL59 | PaTu 8988t | C100(1.62) | LDD1329 | [3] |
| LDCM0451 | CL6 | HEK-293T | C100(0.93); C123(0.94) | LDD1654 | [10] |
| LDCM0452 | CL60 | PaTu 8988t | C100(1.76) | LDD1331 | [3] |
| LDCM0453 | CL61 | HEK-293T | C100(0.91); C123(1.01); C15(1.19) | LDD1656 | [10] |
| LDCM0454 | CL62 | HEK-293T | C100(1.01); C123(1.01) | LDD1657 | [10] |
| LDCM0455 | CL63 | HEK-293T | C100(1.00); C123(1.00) | LDD1658 | [10] |
| LDCM0456 | CL64 | HEK-293T | C100(1.07); C123(0.98) | LDD1659 | [10] |
| LDCM0457 | CL65 | HEK-293T | C100(1.00); C123(1.09) | LDD1660 | [10] |
| LDCM0458 | CL66 | HEK-293T | C100(0.91); C123(0.99) | LDD1661 | [10] |
| LDCM0459 | CL67 | HEK-293T | C100(0.98); C123(1.00) | LDD1662 | [10] |
| LDCM0460 | CL68 | HEK-293T | C100(0.92); C123(1.03) | LDD1663 | [10] |
| LDCM0461 | CL69 | HEK-293T | C100(1.05); C123(1.03) | LDD1664 | [10] |
| LDCM0462 | CL7 | HEK-293T | C100(1.14); C123(0.93) | LDD1665 | [10] |
| LDCM0463 | CL70 | HEK-293T | C100(1.09); C123(1.00) | LDD1666 | [10] |
| LDCM0464 | CL71 | HEK-293T | C100(1.10); C123(1.07) | LDD1667 | [10] |
| LDCM0465 | CL72 | HEK-293T | C100(1.07); C123(1.04) | LDD1668 | [10] |
| LDCM0466 | CL73 | HEK-293T | C100(0.90); C123(1.04); C15(1.09) | LDD1669 | [10] |
| LDCM0467 | CL74 | HEK-293T | C100(1.01); C123(0.98) | LDD1670 | [10] |
| LDCM0469 | CL76 | PaTu 8988t | C100(1.05) | LDD1348 | [3] |
| LDCM0470 | CL77 | PaTu 8988t | C100(0.99) | LDD1349 | [3] |
| LDCM0471 | CL78 | PaTu 8988t | C100(1.47) | LDD1350 | [3] |
| LDCM0472 | CL79 | PaTu 8988t | C100(1.24) | LDD1351 | [3] |
| LDCM0473 | CL8 | HEK-293T | C100(0.88); C123(0.99) | LDD1676 | [10] |
| LDCM0474 | CL80 | PaTu 8988t | C100(1.07) | LDD1353 | [3] |
| LDCM0475 | CL81 | PaTu 8988t | C100(1.25) | LDD1354 | [3] |
| LDCM0476 | CL82 | PaTu 8988t | C100(1.40) | LDD1355 | [3] |
| LDCM0477 | CL83 | PaTu 8988t | C100(1.38) | LDD1356 | [3] |
| LDCM0478 | CL84 | PaTu 8988t | C100(0.92) | LDD1357 | [3] |
| LDCM0479 | CL85 | PaTu 8988t | C100(1.08) | LDD1358 | [3] |
| LDCM0480 | CL86 | PaTu 8988t | C100(1.14) | LDD1359 | [3] |
| LDCM0481 | CL87 | PaTu 8988t | C100(1.27) | LDD1360 | [3] |
| LDCM0482 | CL88 | PaTu 8988t | C100(1.62) | LDD1361 | [3] |
| LDCM0483 | CL89 | PaTu 8988t | C100(1.20) | LDD1362 | [3] |
| LDCM0484 | CL9 | HEK-293T | C100(1.08); C123(0.92) | LDD1687 | [10] |
| LDCM0485 | CL90 | PaTu 8988t | C100(1.34) | LDD1364 | [3] |
| LDCM0486 | CL91 | HEK-293T | C100(1.04); C123(1.01) | LDD1689 | [10] |
| LDCM0487 | CL92 | HEK-293T | C100(1.10); C123(1.00) | LDD1690 | [10] |
| LDCM0488 | CL93 | HEK-293T | C100(1.05); C123(1.03) | LDD1691 | [10] |
| LDCM0489 | CL94 | HEK-293T | C100(1.09); C123(1.00) | LDD1692 | [10] |
| LDCM0490 | CL95 | HEK-293T | C100(1.14); C123(1.04) | LDD1693 | [10] |
| LDCM0491 | CL96 | HEK-293T | C100(0.92); C123(0.97) | LDD1694 | [10] |
| LDCM0492 | CL97 | HEK-293T | C100(0.89); C123(0.97); C15(1.03) | LDD1695 | [10] |
| LDCM0493 | CL98 | HEK-293T | C100(1.09); C123(1.02) | LDD1696 | [10] |
| LDCM0494 | CL99 | HEK-293T | C100(1.08); C123(0.94) | LDD1697 | [10] |
| LDCM0495 | E2913 | HEK-293T | C100(1.11); C123(0.94) | LDD1698 | [10] |
| LDCM0625 | F8 | Ramos | C100(1.05) | LDD2187 | [11] |
| LDCM0574 | Fragment12 | Ramos | C100(1.60) | LDD2191 | [11] |
| LDCM0575 | Fragment13 | Ramos | C100(1.07) | LDD2192 | [11] |
| LDCM0579 | Fragment20 | Ramos | C100(1.53) | LDD2194 | [11] |
| LDCM0580 | Fragment21 | Ramos | C100(2.05) | LDD2195 | [11] |
| LDCM0582 | Fragment23 | Ramos | C100(1.25) | LDD2196 | [11] |
| LDCM0578 | Fragment27 | Ramos | C100(1.46) | LDD2197 | [11] |
| LDCM0586 | Fragment28 | Ramos | C100(0.72) | LDD2198 | [11] |
| LDCM0588 | Fragment30 | Ramos | C100(1.18) | LDD2199 | [11] |
| LDCM0589 | Fragment31 | Ramos | C100(1.36) | LDD2200 | [11] |
| LDCM0590 | Fragment32 | Ramos | C100(3.00) | LDD2201 | [11] |
| LDCM0468 | Fragment33 | HEK-293T | C100(1.01); C123(1.02) | LDD1671 | [10] |
| LDCM0596 | Fragment38 | Ramos | C100(1.58) | LDD2203 | [11] |
| LDCM0427 | Fragment51 | PaTu 8988t | C100(2.50) | LDD1306 | [3] |
| LDCM0610 | Fragment52 | Ramos | C100(1.54) | LDD2204 | [11] |
| LDCM0614 | Fragment56 | Ramos | C100(1.26) | LDD2205 | [11] |
| LDCM0569 | Fragment7 | Ramos | C100(1.38) | LDD2186 | [11] |
| LDCM0571 | Fragment9 | Ramos | C100(1.32) | LDD2188 | [11] |
| LDCM0107 | IAA | HeLa | H66(0.00); H26(0.00); H91(0.00) | LDD0221 | [8] |
| LDCM0022 | KB02 | HCT 116 | C100(3.16) | LDD0080 | [3] |
| LDCM0023 | KB03 | HCT 116 | C100(1.75) | LDD0081 | [3] |
| LDCM0024 | KB05 | HCT 116 | C100(1.45) | LDD0082 | [3] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [8] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Complement component 1 Q subcomponent-binding protein, mitochondrial (C1QBP) | MAM33 family | Q07021 | |||
Transcription factor
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Death domain-associated protein 6 (DAXX) | DAXX family | Q9UER7 | |||
| Nucleophosmin (NPM1) | Nucleoplasmin family | P06748 | |||
References
