Details of the Target
General Information of Target
| Target ID | LDTP07930 | |||||
|---|---|---|---|---|---|---|
| Target Name | Serine/threonine-protein kinase MARK2 (MARK2) | |||||
| Gene Name | MARK2 | |||||
| Gene ID | 2011 | |||||
| Synonyms |
EMK1; Serine/threonine-protein kinase MARK2; EC 2.7.11.1; EC 2.7.11.26; ELKL motif kinase 1; EMK-1; MAP/microtubule affinity-regulating kinase 2; PAR1 homolog; PAR1 homolog b; Par-1b; Par1b |
|||||
| 3D Structure | ||||||
| Sequence |
MSSARTPLPTLNERDTEQPTLGHLDSKPSSKSNMIRGRNSATSADEQPHIGNYRLLKTIG
KGNFAKVKLARHILTGKEVAVKIIDKTQLNSSSLQKLFREVRIMKVLNHPNIVKLFEVIE TEKTLYLVMEYASGGEVFDYLVAHGRMKEKEARAKFRQIVSAVQYCHQKFIVHRDLKAEN LLLDADMNIKIADFGFSNEFTFGNKLDTFCGSPPYAAPELFQGKKYDGPEVDVWSLGVIL YTLVSGSLPFDGQNLKELRERVLRGKYRIPFYMSTDCENLLKKFLILNPSKRGTLEQIMK DRWMNVGHEDDELKPYVEPLPDYKDPRRTELMVSMGYTREEIQDSLVGQRYNEVMATYLL LGYKSSELEGDTITLKPRPSADLTNSSAPSPSHKVQRSVSANPKQRRFSDQAAGPAIPTS NSYSKKTQSNNAENKRPEEDRESGRKASSTAKVPASPLPGLERKKTTPTPSTNSVLSTST NRSRNSPLLERASLGQASIQNGKDSLTMPGSRASTASASAAVSAARPRQHQKSMSASVHP NKASGLPPTESNCEVPRPSTAPQRVPVASPSAHNISSSGGAPDRTNFPRGVSSRSTFHAG QLRQVRDQQNLPYGVTPASPSGHSQGRRGASGSIFSKFTSKFVRRNLSFRFARRNLNEPE SKDRVETLRPHVVGSGGNDKEKEEFREAKPRSLRFTWSMKTTSSMEPNEMMREIRKVLDA NSCQSELHEKYMLLCMHGTPGHEDFVQWEMEVCKLPRLSLNGVRFKRISGTSMAFKNIAS KIANELKL |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Protein kinase superfamily, CAMK Ser/Thr protein kinase family, SNF1 subfamily
|
|||||
| Subcellular location |
Cell membrane
|
|||||
| Function |
Serine/threonine-protein kinase. Involved in cell polarity and microtubule dynamics regulation. Phosphorylates CRTC2/TORC2, DCX, HDAC7, KIF13B, MAP2, MAP4 and RAB11FIP2. Phosphorylates the microtubule-associated protein MAPT/TAU. Plays a key role in cell polarity by phosphorylating the microtubule-associated proteins MAP2, MAP4 and MAPT/TAU at KXGS motifs, causing detachment from microtubules, and their disassembly. Regulates epithelial cell polarity by phosphorylating RAB11FIP2. Involved in the regulation of neuronal migration through its dual activities in regulating cellular polarity and microtubule dynamics, possibly by phosphorylating and regulating DCX. Regulates axogenesis by phosphorylating KIF13B, promoting interaction between KIF13B and 14-3-3 and inhibiting microtubule-dependent accumulation of KIF13B. Also required for neurite outgrowth and establishment of neuronal polarity. Regulates localization and activity of some histone deacetylases by mediating phosphorylation of HDAC7, promoting subsequent interaction between HDAC7 and 14-3-3 and export from the nucleus. Also acts as a positive regulator of the Wnt signaling pathway, probably by mediating phosphorylation of dishevelled proteins (DVL1, DVL2 and/or DVL3). Modulates the developmental decision to build a columnar versus a hepatic epithelial cell apparently by promoting a switch from a direct to a transcytotic mode of apical protein delivery. Essential for the asymmetric development of membrane domains of polarized epithelial cells.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| DU145 | SNV: p.R259P | DBIA Probe Info | |||
| HDMYZ | SNV: p.K464R | DBIA Probe Info | |||
| HT115 | SNV: p.R407H | DBIA Probe Info | |||
| KYSE150 | Substitution: p.P588F | DBIA Probe Info | |||
| KYSE30 | Substitution: p.A217S | DBIA Probe Info | |||
| LNCaP clone FGC | SNV: p.L21S | DBIA Probe Info | |||
| MELHO | Substitution: p.R445Q | DBIA Probe Info | |||
| MEWO | SNV: p.R153G | DBIA Probe Info | |||
| OVK18 | Insertion: p.Y226IfsTer2 | DBIA Probe Info | |||
| RKO | SNV: p.A70T | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
14.52 | LDD0402 | [1] | |
|
TH211 Probe Info |
 |
Y53(9.07) | LDD0257 | [2] | |
|
STPyne Probe Info |
 |
K641(5.77); K776(9.20) | LDD0277 | [3] | |
|
BTD Probe Info |
 |
C723(0.83) | LDD2089 | [4] | |
|
DBIA Probe Info |
 |
C553(13.77) | LDD0204 | [5] | |
|
IPM Probe Info |
 |
C723(1.77) | LDD1702 | [4] | |
|
ATP probe Probe Info |
 |
K96(0.00); K394(0.00) | LDD0199 | [6] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [7] | |
|
Lodoacetamide azide Probe Info |
 |
C166(0.00); C723(0.00); C277(0.00) | LDD0037 | [7] | |
|
ATP probe Probe Info |
 |
K177(0.00); K82(0.00) | LDD0035 | [8] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [9] | |
|
KY-26 Probe Info |
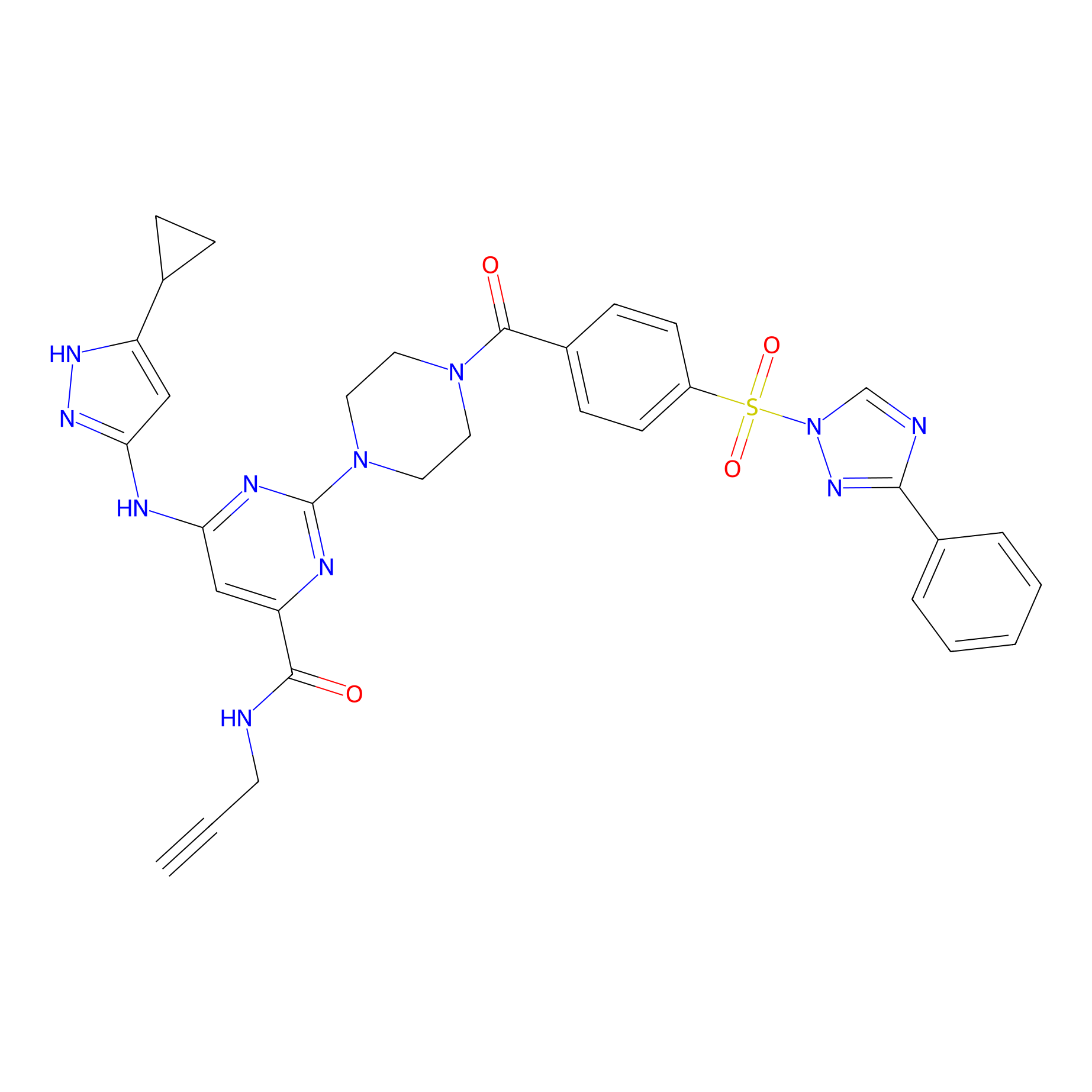 |
N.A. | LDD0301 | [10] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [11] | |
|
AOyne Probe Info |
 |
6.30 | LDD0443 | [12] | |
|
NAIA_5 Probe Info |
 |
C723(0.00); C277(0.00) | LDD2223 | [13] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C338 Probe Info |
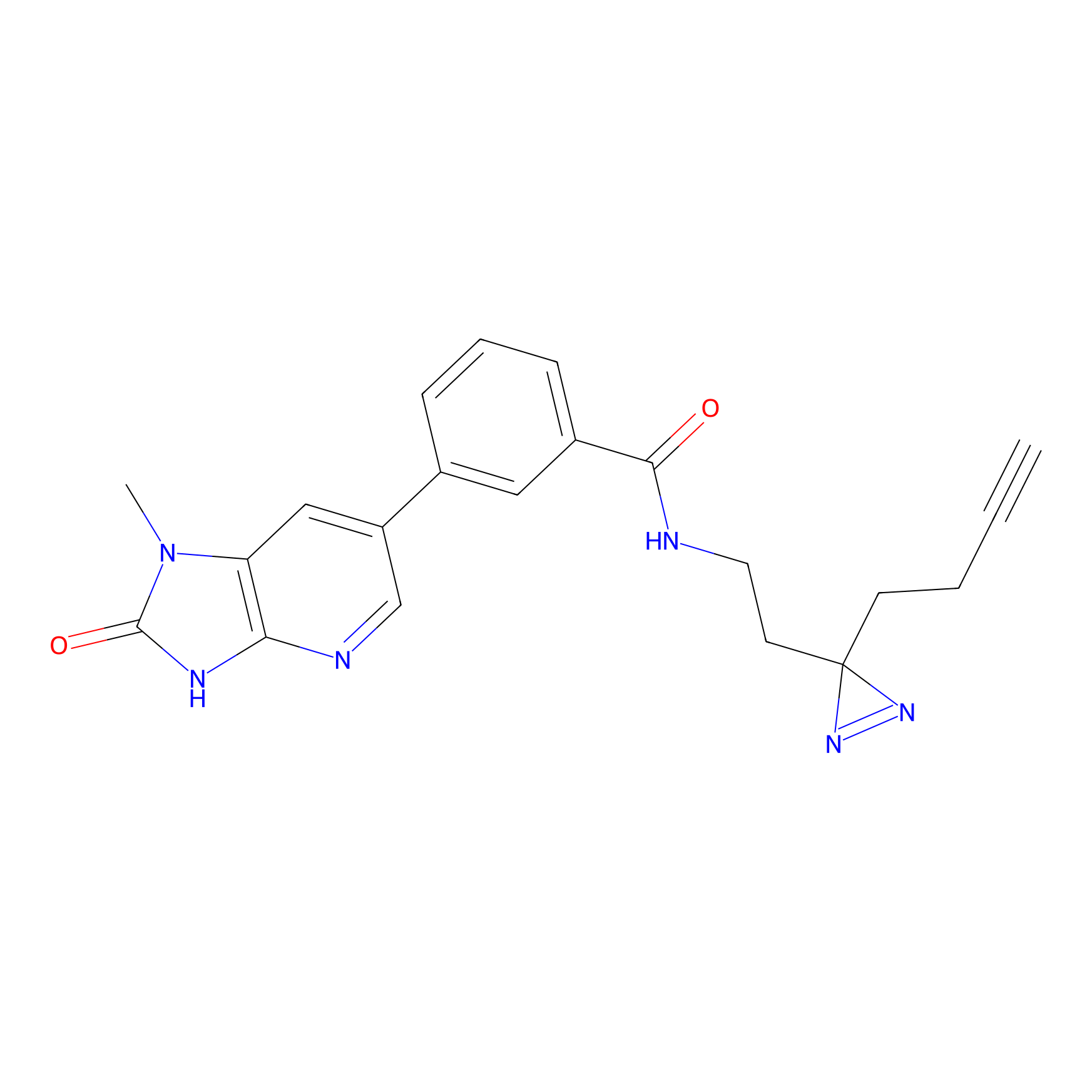 |
17.03 | LDD2001 | [14] | |
|
Staurosporine capture compound Probe Info |
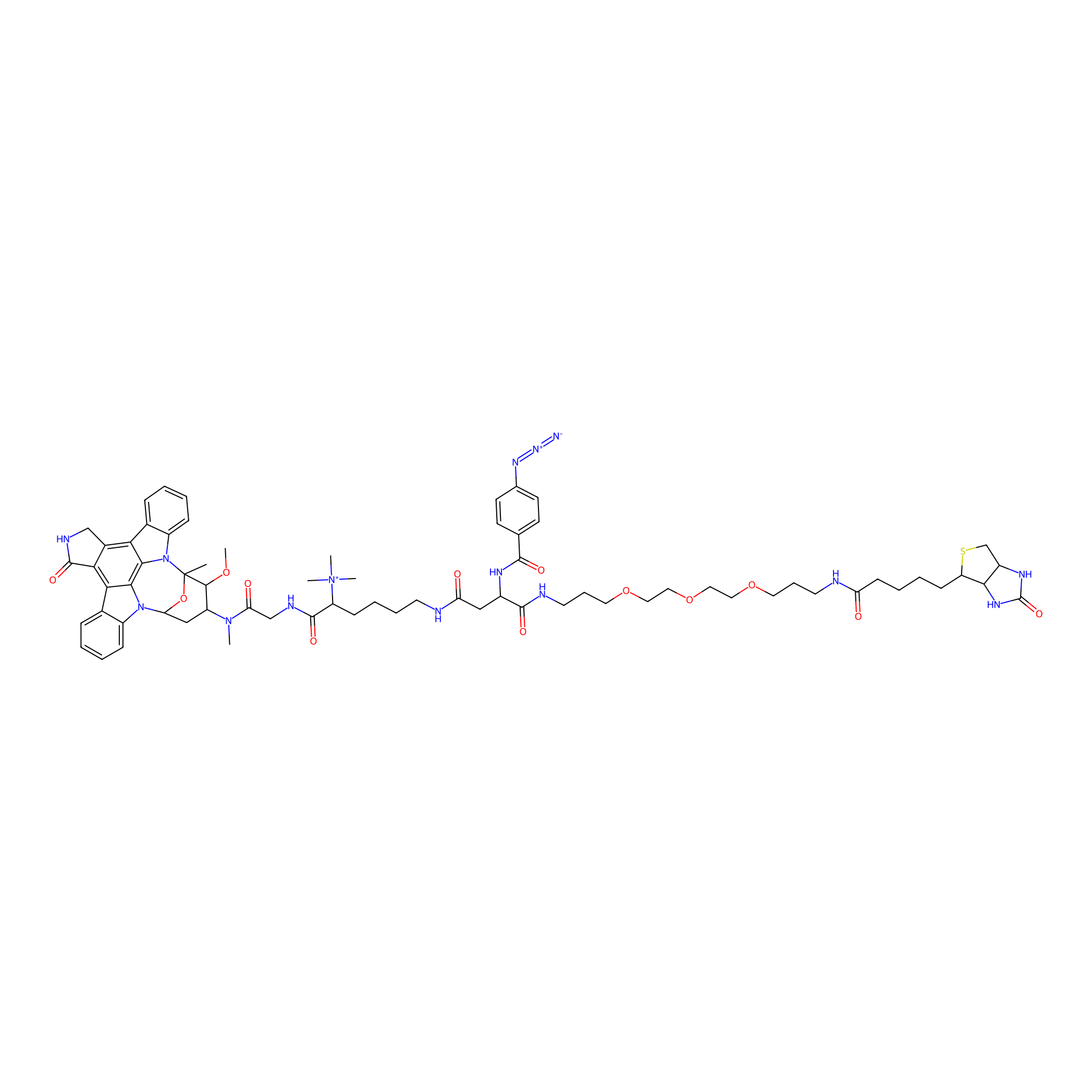 |
N.A. | LDD0083 | [15] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0537 | 2-Cyano-N,N-dimethylacetamide | MDA-MB-231 | C723(0.65) | LDD2130 | [4] |
| LDCM0214 | AC1 | HEK-293T | C553(1.19) | LDD1507 | [16] |
| LDCM0215 | AC10 | HEK-293T | C553(0.89) | LDD1508 | [16] |
| LDCM0226 | AC11 | HEK-293T | C553(1.01) | LDD1509 | [16] |
| LDCM0237 | AC12 | HEK-293T | C553(1.17) | LDD1510 | [16] |
| LDCM0259 | AC14 | HEK-293T | C553(0.86) | LDD1512 | [16] |
| LDCM0270 | AC15 | HEK-293T | C553(0.91) | LDD1513 | [16] |
| LDCM0276 | AC17 | HEK-293T | C553(1.04) | LDD1515 | [16] |
| LDCM0277 | AC18 | HEK-293T | C553(0.99) | LDD1516 | [16] |
| LDCM0278 | AC19 | HEK-293T | C553(1.16) | LDD1517 | [16] |
| LDCM0279 | AC2 | HEK-293T | C553(0.94) | LDD1518 | [16] |
| LDCM0280 | AC20 | HEK-293T | C553(1.08) | LDD1519 | [16] |
| LDCM0281 | AC21 | HEK-293T | C553(1.03) | LDD1520 | [16] |
| LDCM0282 | AC22 | HEK-293T | C553(0.83) | LDD1521 | [16] |
| LDCM0283 | AC23 | HEK-293T | C553(0.89) | LDD1522 | [16] |
| LDCM0285 | AC25 | HEK-293T | C553(1.09) | LDD1524 | [16] |
| LDCM0286 | AC26 | HEK-293T | C553(0.97) | LDD1525 | [16] |
| LDCM0287 | AC27 | HEK-293T | C553(1.16) | LDD1526 | [16] |
| LDCM0288 | AC28 | HEK-293T | C553(1.03) | LDD1527 | [16] |
| LDCM0289 | AC29 | HEK-293T | C553(1.01) | LDD1528 | [16] |
| LDCM0290 | AC3 | HEK-293T | C553(0.99) | LDD1529 | [16] |
| LDCM0291 | AC30 | HEK-293T | C553(1.06) | LDD1530 | [16] |
| LDCM0292 | AC31 | HEK-293T | C553(0.93) | LDD1531 | [16] |
| LDCM0294 | AC33 | HEK-293T | C553(1.22) | LDD1533 | [16] |
| LDCM0295 | AC34 | HEK-293T | C553(0.87) | LDD1534 | [16] |
| LDCM0296 | AC35 | HEK-293T | C553(1.00) | LDD1535 | [16] |
| LDCM0297 | AC36 | HEK-293T | C553(0.84) | LDD1536 | [16] |
| LDCM0298 | AC37 | HEK-293T | C553(1.05) | LDD1537 | [16] |
| LDCM0299 | AC38 | HEK-293T | C553(0.83) | LDD1538 | [16] |
| LDCM0300 | AC39 | HEK-293T | C553(0.91) | LDD1539 | [16] |
| LDCM0301 | AC4 | HEK-293T | C553(0.95) | LDD1540 | [16] |
| LDCM0303 | AC41 | HEK-293T | C553(1.13) | LDD1542 | [16] |
| LDCM0304 | AC42 | HEK-293T | C553(1.03) | LDD1543 | [16] |
| LDCM0305 | AC43 | HEK-293T | C553(1.11) | LDD1544 | [16] |
| LDCM0306 | AC44 | HEK-293T | C553(1.01) | LDD1545 | [16] |
| LDCM0307 | AC45 | HEK-293T | C553(1.09) | LDD1546 | [16] |
| LDCM0308 | AC46 | HEK-293T | C553(1.04) | LDD1547 | [16] |
| LDCM0309 | AC47 | HEK-293T | C553(0.87) | LDD1548 | [16] |
| LDCM0311 | AC49 | HEK-293T | C553(1.12) | LDD1550 | [16] |
| LDCM0312 | AC5 | HEK-293T | C553(1.04) | LDD1551 | [16] |
| LDCM0313 | AC50 | HEK-293T | C553(1.05) | LDD1552 | [16] |
| LDCM0314 | AC51 | HEK-293T | C553(1.10) | LDD1553 | [16] |
| LDCM0315 | AC52 | HEK-293T | C553(1.18) | LDD1554 | [16] |
| LDCM0316 | AC53 | HEK-293T | C553(1.06) | LDD1555 | [16] |
| LDCM0317 | AC54 | HEK-293T | C553(0.95) | LDD1556 | [16] |
| LDCM0318 | AC55 | HEK-293T | C553(0.86) | LDD1557 | [16] |
| LDCM0320 | AC57 | HEK-293T | C553(1.22) | LDD1559 | [16] |
| LDCM0321 | AC58 | HEK-293T | C553(0.88) | LDD1560 | [16] |
| LDCM0322 | AC59 | HEK-293T | C553(1.04) | LDD1561 | [16] |
| LDCM0323 | AC6 | HEK-293T | C553(0.83) | LDD1562 | [16] |
| LDCM0324 | AC60 | HEK-293T | C553(1.02) | LDD1563 | [16] |
| LDCM0325 | AC61 | HEK-293T | C553(1.09) | LDD1564 | [16] |
| LDCM0326 | AC62 | HEK-293T | C553(0.88) | LDD1565 | [16] |
| LDCM0327 | AC63 | HEK-293T | C553(1.08) | LDD1566 | [16] |
| LDCM0334 | AC7 | HEK-293T | C553(1.01) | LDD1568 | [16] |
| LDCM0248 | AKOS034007472 | HEK-293T | C553(1.01) | LDD1511 | [16] |
| LDCM0356 | AKOS034007680 | HEK-293T | C553(1.09) | LDD1570 | [16] |
| LDCM0156 | Aniline | NCI-H1299 | 14.10 | LDD0403 | [1] |
| LDCM0102 | BDHI 8 | Jurkat | C553(13.77) | LDD0204 | [5] |
| LDCM0632 | CL-Sc | Hep-G2 | C166(3.40) | LDD2227 | [13] |
| LDCM0367 | CL1 | HEK-293T | C553(1.04); C723(1.11) | LDD1571 | [16] |
| LDCM0368 | CL10 | HEK-293T | C553(1.01) | LDD1572 | [16] |
| LDCM0369 | CL100 | HEK-293T | C553(1.08); C723(1.02) | LDD1573 | [16] |
| LDCM0370 | CL101 | HEK-293T | C553(1.17); C723(1.03) | LDD1574 | [16] |
| LDCM0372 | CL103 | HEK-293T | C553(1.02) | LDD1576 | [16] |
| LDCM0373 | CL104 | HEK-293T | C553(1.09); C723(0.99) | LDD1577 | [16] |
| LDCM0374 | CL105 | HEK-293T | C553(0.96); C723(1.03) | LDD1578 | [16] |
| LDCM0376 | CL107 | HEK-293T | C553(0.94) | LDD1580 | [16] |
| LDCM0377 | CL108 | HEK-293T | C553(0.91); C723(0.95) | LDD1581 | [16] |
| LDCM0378 | CL109 | HEK-293T | C553(0.99); C723(0.95) | LDD1582 | [16] |
| LDCM0379 | CL11 | HEK-293T | C553(0.93) | LDD1583 | [16] |
| LDCM0381 | CL111 | HEK-293T | C553(0.98) | LDD1585 | [16] |
| LDCM0382 | CL112 | HEK-293T | C553(1.05); C723(0.98) | LDD1586 | [16] |
| LDCM0383 | CL113 | HEK-293T | C553(0.91); C723(0.90) | LDD1587 | [16] |
| LDCM0385 | CL115 | HEK-293T | C553(0.99) | LDD1589 | [16] |
| LDCM0386 | CL116 | HEK-293T | C553(1.12); C723(1.06) | LDD1590 | [16] |
| LDCM0387 | CL117 | HEK-293T | C553(1.00); C723(1.00) | LDD1591 | [16] |
| LDCM0389 | CL119 | HEK-293T | C553(1.07) | LDD1593 | [16] |
| LDCM0391 | CL120 | HEK-293T | C553(1.05); C723(1.02) | LDD1595 | [16] |
| LDCM0392 | CL121 | HEK-293T | C553(0.90); C723(0.97) | LDD1596 | [16] |
| LDCM0394 | CL123 | HEK-293T | C553(0.82) | LDD1598 | [16] |
| LDCM0395 | CL124 | HEK-293T | C553(1.13); C723(1.00) | LDD1599 | [16] |
| LDCM0396 | CL125 | HEK-293T | C553(0.86); C723(0.81) | LDD1600 | [16] |
| LDCM0398 | CL127 | HEK-293T | C553(1.06) | LDD1602 | [16] |
| LDCM0399 | CL128 | HEK-293T | C553(1.32); C723(1.00) | LDD1603 | [16] |
| LDCM0400 | CL13 | HEK-293T | C553(1.24); C723(0.97) | LDD1604 | [16] |
| LDCM0402 | CL15 | HEK-293T | C553(1.05) | LDD1606 | [16] |
| LDCM0403 | CL16 | HEK-293T | C553(1.04); C723(1.15) | LDD1607 | [16] |
| LDCM0404 | CL17 | HEK-293T | C553(0.97) | LDD1608 | [16] |
| LDCM0405 | CL18 | HEK-293T | C553(1.02) | LDD1609 | [16] |
| LDCM0406 | CL19 | HEK-293T | C553(1.15) | LDD1610 | [16] |
| LDCM0408 | CL20 | HEK-293T | C553(1.27) | LDD1612 | [16] |
| LDCM0409 | CL21 | HEK-293T | C553(0.88) | LDD1613 | [16] |
| LDCM0410 | CL22 | HEK-293T | C553(1.04) | LDD1614 | [16] |
| LDCM0411 | CL23 | HEK-293T | C553(0.98) | LDD1615 | [16] |
| LDCM0413 | CL25 | HEK-293T | C553(0.89); C723(0.92) | LDD1617 | [16] |
| LDCM0415 | CL27 | HEK-293T | C553(1.07) | LDD1619 | [16] |
| LDCM0416 | CL28 | HEK-293T | C553(1.04); C723(0.99) | LDD1620 | [16] |
| LDCM0417 | CL29 | HEK-293T | C553(1.04) | LDD1621 | [16] |
| LDCM0418 | CL3 | HEK-293T | C553(0.84) | LDD1622 | [16] |
| LDCM0419 | CL30 | HEK-293T | C553(0.87) | LDD1623 | [16] |
| LDCM0420 | CL31 | HEK-293T | C553(0.89) | LDD1624 | [16] |
| LDCM0421 | CL32 | HEK-293T | C553(1.19) | LDD1625 | [16] |
| LDCM0422 | CL33 | HEK-293T | C553(0.82) | LDD1626 | [16] |
| LDCM0423 | CL34 | HEK-293T | C553(1.03) | LDD1627 | [16] |
| LDCM0424 | CL35 | HEK-293T | C553(0.78) | LDD1628 | [16] |
| LDCM0426 | CL37 | HEK-293T | C553(0.97); C723(1.24) | LDD1630 | [16] |
| LDCM0428 | CL39 | HEK-293T | C553(0.93) | LDD1632 | [16] |
| LDCM0429 | CL4 | HEK-293T | C553(1.16); C723(1.06) | LDD1633 | [16] |
| LDCM0430 | CL40 | HEK-293T | C553(0.99); C723(0.96) | LDD1634 | [16] |
| LDCM0431 | CL41 | HEK-293T | C553(1.08) | LDD1635 | [16] |
| LDCM0432 | CL42 | HEK-293T | C553(1.05) | LDD1636 | [16] |
| LDCM0433 | CL43 | HEK-293T | C553(1.11) | LDD1637 | [16] |
| LDCM0434 | CL44 | HEK-293T | C553(1.01) | LDD1638 | [16] |
| LDCM0435 | CL45 | HEK-293T | C553(0.87) | LDD1639 | [16] |
| LDCM0436 | CL46 | HEK-293T | C553(1.02) | LDD1640 | [16] |
| LDCM0437 | CL47 | HEK-293T | C553(0.81) | LDD1641 | [16] |
| LDCM0439 | CL49 | HEK-293T | C553(1.04); C723(0.93) | LDD1643 | [16] |
| LDCM0440 | CL5 | HEK-293T | C553(1.17) | LDD1644 | [16] |
| LDCM0443 | CL52 | HEK-293T | C553(0.92); C723(1.10) | LDD1646 | [16] |
| LDCM0444 | CL53 | HEK-293T | C553(1.10) | LDD1647 | [16] |
| LDCM0445 | CL54 | HEK-293T | C553(0.96) | LDD1648 | [16] |
| LDCM0446 | CL55 | HEK-293T | C553(1.06) | LDD1649 | [16] |
| LDCM0447 | CL56 | HEK-293T | C553(1.17) | LDD1650 | [16] |
| LDCM0448 | CL57 | HEK-293T | C553(0.90) | LDD1651 | [16] |
| LDCM0449 | CL58 | HEK-293T | C553(0.88) | LDD1652 | [16] |
| LDCM0450 | CL59 | HEK-293T | C553(0.78) | LDD1653 | [16] |
| LDCM0451 | CL6 | HEK-293T | C553(1.04) | LDD1654 | [16] |
| LDCM0453 | CL61 | HEK-293T | C553(0.86); C723(1.10) | LDD1656 | [16] |
| LDCM0455 | CL63 | HEK-293T | C553(0.99) | LDD1658 | [16] |
| LDCM0456 | CL64 | HEK-293T | C553(1.16); C723(1.08) | LDD1659 | [16] |
| LDCM0457 | CL65 | HEK-293T | C553(1.04) | LDD1660 | [16] |
| LDCM0458 | CL66 | HEK-293T | C553(0.88) | LDD1661 | [16] |
| LDCM0459 | CL67 | HEK-293T | C553(1.07) | LDD1662 | [16] |
| LDCM0460 | CL68 | HEK-293T | C553(1.15) | LDD1663 | [16] |
| LDCM0461 | CL69 | HEK-293T | C553(0.97) | LDD1664 | [16] |
| LDCM0462 | CL7 | HEK-293T | C553(1.12) | LDD1665 | [16] |
| LDCM0463 | CL70 | HEK-293T | C553(1.04) | LDD1666 | [16] |
| LDCM0464 | CL71 | HEK-293T | C553(0.90) | LDD1667 | [16] |
| LDCM0466 | CL73 | HEK-293T | C553(0.94); C723(1.14) | LDD1669 | [16] |
| LDCM0469 | CL76 | HEK-293T | C553(1.06); C723(1.06) | LDD1672 | [16] |
| LDCM0470 | CL77 | HEK-293T | C553(1.08) | LDD1673 | [16] |
| LDCM0471 | CL78 | HEK-293T | C553(0.96) | LDD1674 | [16] |
| LDCM0472 | CL79 | HEK-293T | C553(1.05) | LDD1675 | [16] |
| LDCM0473 | CL8 | HEK-293T | C553(0.69) | LDD1676 | [16] |
| LDCM0474 | CL80 | HEK-293T | C553(1.70) | LDD1677 | [16] |
| LDCM0475 | CL81 | HEK-293T | C553(0.88) | LDD1678 | [16] |
| LDCM0476 | CL82 | HEK-293T | C553(0.92) | LDD1679 | [16] |
| LDCM0477 | CL83 | HEK-293T | C553(0.91) | LDD1680 | [16] |
| LDCM0479 | CL85 | HEK-293T | C553(0.94); C723(0.95) | LDD1682 | [16] |
| LDCM0481 | CL87 | HEK-293T | C553(1.06) | LDD1684 | [16] |
| LDCM0482 | CL88 | HEK-293T | C553(1.27); C723(0.99) | LDD1685 | [16] |
| LDCM0483 | CL89 | HEK-293T | C553(1.31) | LDD1686 | [16] |
| LDCM0484 | CL9 | HEK-293T | C553(1.00) | LDD1687 | [16] |
| LDCM0485 | CL90 | HEK-293T | C553(1.01) | LDD1688 | [16] |
| LDCM0486 | CL91 | HEK-293T | C553(1.08) | LDD1689 | [16] |
| LDCM0487 | CL92 | HEK-293T | C553(1.20) | LDD1690 | [16] |
| LDCM0488 | CL93 | HEK-293T | C553(0.91) | LDD1691 | [16] |
| LDCM0489 | CL94 | HEK-293T | C553(0.97) | LDD1692 | [16] |
| LDCM0490 | CL95 | HEK-293T | C553(0.88) | LDD1693 | [16] |
| LDCM0492 | CL97 | HEK-293T | C553(0.93); C723(0.91) | LDD1695 | [16] |
| LDCM0494 | CL99 | HEK-293T | C553(1.08) | LDD1697 | [16] |
| LDCM0495 | E2913 | HEK-293T | C553(1.00) | LDD1698 | [16] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C723(1.77) | LDD1702 | [4] |
| LDCM0468 | Fragment33 | HEK-293T | C553(1.07) | LDD1671 | [16] |
| LDCM0022 | KB02 | HEK-293T | C553(1.33) | LDD1492 | [16] |
| LDCM0023 | KB03 | Jurkat | C277(83.81); C210(5.76) | LDD0209 | [5] |
| LDCM0024 | KB05 | G361 | C82(3.31); C38(2.21) | LDD3311 | [17] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C723(0.64) | LDD2121 | [4] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C723(0.83) | LDD2089 | [4] |
| LDCM0019 | Staurosporine | Hep-G2 | N.A. | LDD0083 | [15] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| 14-3-3 protein gamma (YWHAG) | 14-3-3 family | P61981 | |||
| 14-3-3 protein zeta/delta (YWHAZ) | 14-3-3 family | P63104 | |||
Other
References
