Details of the Target
General Information of Target
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| CHL1 | SNV: p.V297I | DBIA Probe Info | |||
| MOLT4 | SNV: p.T264I | IA-alkyne Probe Info | |||
| NB1 | Deletion: p.E322DfsTer17; p.E322SfsTer17 | . | |||
| YSCCC | SNV: p.E83D; p.M114I | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TH211 Probe Info |
 |
Y163(9.10); Y150(8.13); Y67(6.67); Y32(6.37) | LDD0257 | [1] | |
|
m-APA Probe Info |
 |
11.55 | LDD0403 | [2] | |
|
Alkylaryl probe 2 Probe Info |
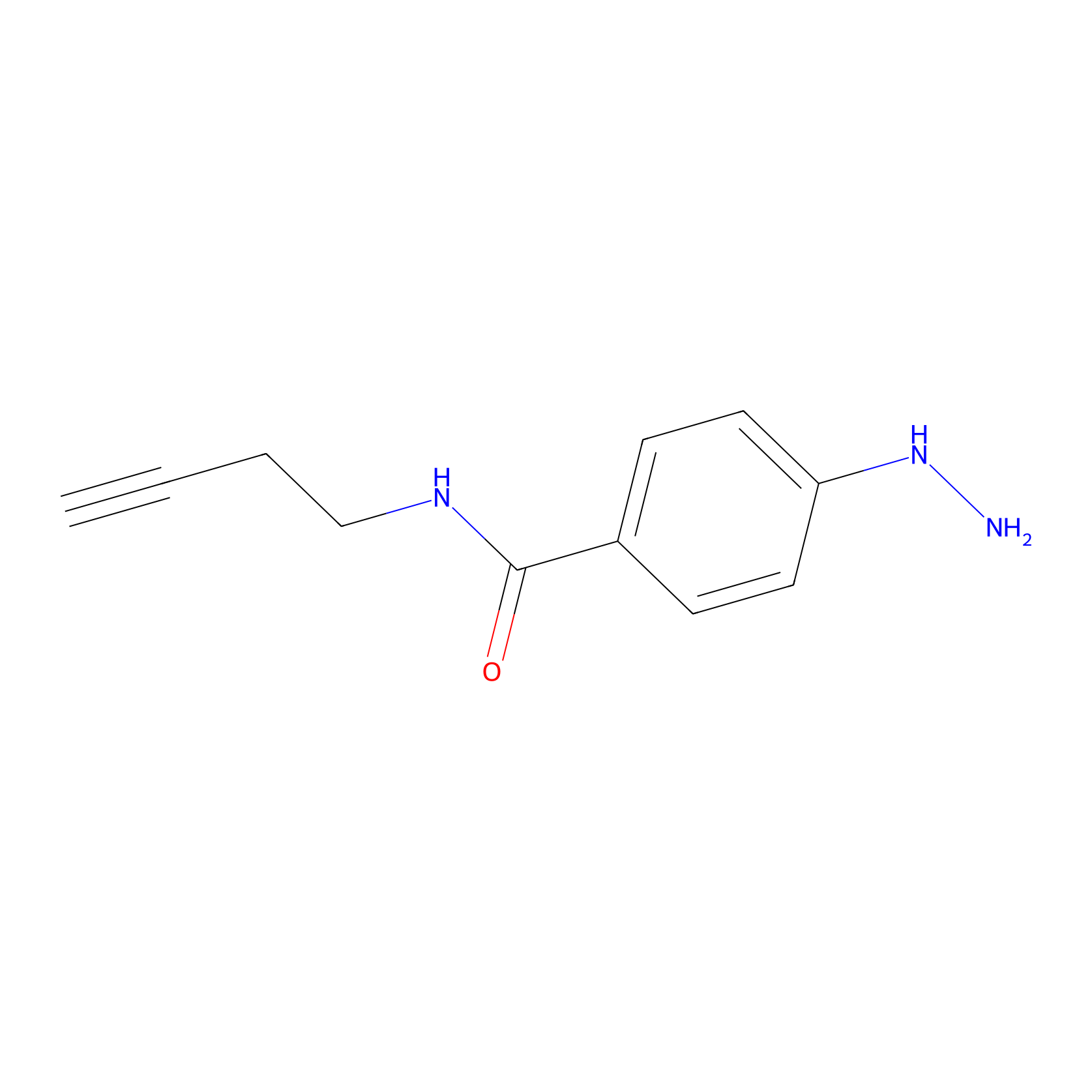 |
3.00 | LDD0390 | [3] | |
|
DBIA Probe Info |
 |
C207(3.42) | LDD3310 | [4] | |
|
BTD Probe Info |
 |
C101(1.07) | LDD2120 | [5] | |
|
HHS-475 Probe Info |
 |
Y32(1.48) | LDD0264 | [6] | |
|
ATP probe Probe Info |
 |
K167(0.00); K142(0.00) | LDD0199 | [7] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C101(0.00); C207(0.00) | LDD0038 | [8] | |
|
IA-alkyne Probe Info |
 |
C101(0.00); C207(0.00) | LDD0036 | [8] | |
|
IPIAA_L Probe Info |
 |
N.A. | LDD0031 | [9] | |
|
Lodoacetamide azide Probe Info |
 |
C101(0.00); C207(0.00) | LDD0037 | [8] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [10] | |
|
IPM Probe Info |
 |
C380(0.00); C394(0.00) | LDD2156 | [11] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [12] | |
|
AOyne Probe Info |
 |
6.50 | LDD0443 | [13] | |
|
HHS-482 Probe Info |
 |
Y32(0.88) | LDD2239 | [14] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C101(2.01) | LDD0372 | [15] |
| LDCM0237 | AC12 | HEK-293T | C101(1.05) | LDD1510 | [16] |
| LDCM0259 | AC14 | HEK-293T | C101(1.04) | LDD1512 | [16] |
| LDCM0280 | AC20 | HEK-293T | C101(0.90) | LDD1519 | [16] |
| LDCM0282 | AC22 | HEK-293T | C101(0.76) | LDD1521 | [16] |
| LDCM0284 | AC24 | HEK-293T | C101(1.27) | LDD1523 | [16] |
| LDCM0288 | AC28 | HEK-293T | C101(0.97) | LDD1527 | [16] |
| LDCM0291 | AC30 | HEK-293T | C101(0.98) | LDD1530 | [16] |
| LDCM0293 | AC32 | HEK-293T | C101(1.16) | LDD1532 | [16] |
| LDCM0297 | AC36 | HEK-293T | C101(0.89) | LDD1536 | [16] |
| LDCM0299 | AC38 | HEK-293T | C101(1.05) | LDD1538 | [16] |
| LDCM0301 | AC4 | HEK-293T | C101(0.90) | LDD1540 | [16] |
| LDCM0302 | AC40 | HEK-293T | C101(1.07) | LDD1541 | [16] |
| LDCM0306 | AC44 | HEK-293T | C101(1.07) | LDD1545 | [16] |
| LDCM0308 | AC46 | HEK-293T | C101(1.07) | LDD1547 | [16] |
| LDCM0310 | AC48 | HEK-293T | C101(1.36) | LDD1549 | [16] |
| LDCM0315 | AC52 | HEK-293T | C101(1.07) | LDD1554 | [16] |
| LDCM0317 | AC54 | HEK-293T | C101(1.01) | LDD1556 | [16] |
| LDCM0319 | AC56 | HEK-293T | C101(1.07) | LDD1558 | [16] |
| LDCM0323 | AC6 | HEK-293T | C101(0.96) | LDD1562 | [16] |
| LDCM0324 | AC60 | HEK-293T | C101(0.94) | LDD1563 | [16] |
| LDCM0326 | AC62 | HEK-293T | C101(1.05) | LDD1565 | [16] |
| LDCM0328 | AC64 | HEK-293T | C101(1.35) | LDD1567 | [16] |
| LDCM0345 | AC8 | HEK-293T | C101(1.38) | LDD1569 | [16] |
| LDCM0275 | AKOS034007705 | HEK-293T | C101(1.02) | LDD1514 | [16] |
| LDCM0156 | Aniline | NCI-H1299 | 11.55 | LDD0403 | [2] |
| LDCM0367 | CL1 | HEK-293T | C101(0.83) | LDD1571 | [16] |
| LDCM0368 | CL10 | HEK-293T | C101(0.93) | LDD1572 | [16] |
| LDCM0370 | CL101 | HEK-293T | C101(0.94) | LDD1574 | [16] |
| LDCM0374 | CL105 | HEK-293T | C101(1.03) | LDD1578 | [16] |
| LDCM0378 | CL109 | HEK-293T | C101(1.15) | LDD1582 | [16] |
| LDCM0383 | CL113 | HEK-293T | C101(0.91) | LDD1587 | [16] |
| LDCM0387 | CL117 | HEK-293T | C101(1.00) | LDD1591 | [16] |
| LDCM0390 | CL12 | HEK-293T | C101(1.46) | LDD1594 | [16] |
| LDCM0392 | CL121 | HEK-293T | C101(1.01) | LDD1596 | [16] |
| LDCM0396 | CL125 | HEK-293T | C101(1.19) | LDD1600 | [16] |
| LDCM0400 | CL13 | HEK-293T | C101(0.94) | LDD1604 | [16] |
| LDCM0408 | CL20 | HEK-293T | C101(1.09) | LDD1612 | [16] |
| LDCM0410 | CL22 | HEK-293T | C101(1.16) | LDD1614 | [16] |
| LDCM0412 | CL24 | HEK-293T | C101(1.31) | LDD1616 | [16] |
| LDCM0413 | CL25 | HEK-293T | C101(0.93) | LDD1617 | [16] |
| LDCM0421 | CL32 | HEK-293T | C101(1.01) | LDD1625 | [16] |
| LDCM0423 | CL34 | HEK-293T | C101(1.14) | LDD1627 | [16] |
| LDCM0425 | CL36 | HEK-293T | C101(1.30) | LDD1629 | [16] |
| LDCM0426 | CL37 | HEK-293T | C101(1.03) | LDD1630 | [16] |
| LDCM0434 | CL44 | HEK-293T | C101(1.13) | LDD1638 | [16] |
| LDCM0436 | CL46 | HEK-293T | C101(1.11) | LDD1640 | [16] |
| LDCM0438 | CL48 | HEK-293T | C101(1.04) | LDD1642 | [16] |
| LDCM0439 | CL49 | HEK-293T | C101(1.18) | LDD1643 | [16] |
| LDCM0447 | CL56 | HEK-293T | C101(1.04) | LDD1650 | [16] |
| LDCM0449 | CL58 | HEK-293T | C101(1.20) | LDD1652 | [16] |
| LDCM0452 | CL60 | HEK-293T | C101(1.10) | LDD1655 | [16] |
| LDCM0453 | CL61 | HEK-293T | C101(0.88) | LDD1656 | [16] |
| LDCM0460 | CL68 | HEK-293T | C101(1.21) | LDD1663 | [16] |
| LDCM0463 | CL70 | HEK-293T | C101(1.26) | LDD1666 | [16] |
| LDCM0465 | CL72 | HEK-293T | C101(1.16) | LDD1668 | [16] |
| LDCM0466 | CL73 | HEK-293T | C101(0.94) | LDD1669 | [16] |
| LDCM0473 | CL8 | HEK-293T | C101(1.12) | LDD1676 | [16] |
| LDCM0474 | CL80 | HEK-293T | C101(1.07) | LDD1677 | [16] |
| LDCM0476 | CL82 | HEK-293T | C101(1.01) | LDD1679 | [16] |
| LDCM0478 | CL84 | HEK-293T | C101(1.01) | LDD1681 | [16] |
| LDCM0479 | CL85 | HEK-293T | C101(0.86) | LDD1682 | [16] |
| LDCM0487 | CL92 | HEK-293T | C101(0.99) | LDD1690 | [16] |
| LDCM0489 | CL94 | HEK-293T | C101(0.96) | LDD1692 | [16] |
| LDCM0491 | CL96 | HEK-293T | C101(1.13) | LDD1694 | [16] |
| LDCM0492 | CL97 | HEK-293T | C101(0.90) | LDD1695 | [16] |
| LDCM0116 | HHS-0101 | DM93 | Y32(1.48) | LDD0264 | [6] |
| LDCM0117 | HHS-0201 | DM93 | Y32(1.70) | LDD0265 | [6] |
| LDCM0118 | HHS-0301 | DM93 | Y32(2.34) | LDD0266 | [6] |
| LDCM0119 | HHS-0401 | DM93 | Y32(1.64) | LDD0267 | [6] |
| LDCM0120 | HHS-0701 | DM93 | Y32(1.53) | LDD0268 | [6] |
| LDCM0022 | KB02 | HEK-293T | C101(1.00); C207(0.76) | LDD1492 | [16] |
| LDCM0023 | KB03 | HEK-293T | C101(1.11); C207(0.95) | LDD1497 | [16] |
| LDCM0024 | KB05 | COLO792 | C207(3.42) | LDD3310 | [4] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C101(1.07) | LDD2120 | [5] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C101(1.07) | LDD2128 | [5] |
| LDCM0551 | Nucleophilic fragment 5b | MDA-MB-231 | C101(0.20) | LDD2145 | [5] |
| LDCM0099 | Phenelzine | HEK-293T | 3.00 | LDD0390 | [3] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Cellular tumor antigen p53 (TP53) | P53 family | P04637 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Estrogen receptor (ESR1) | Nuclear hormone receptor family | P03372 | |||
Other
References
