Details of the Target
General Information of Target
| Target ID | LDTP07053 | |||||
|---|---|---|---|---|---|---|
| Target Name | Cytochrome c oxidase assembly protein COX20, mitochondrial (COX20) | |||||
| Gene Name | COX20 | |||||
| Gene ID | 116228 | |||||
| Synonyms |
FAM36A; Cytochrome c oxidase assembly protein COX20, mitochondrial |
|||||
| 3D Structure | ||||||
| Sequence |
MAAPPEPGEPEERKSLKLLGFLDVENTPCARHSILYGSLGSVVAGFGHFLFTSRIRRSCD
VGVGGFILVTLGCWFHCRYNYAKQRIQERIAREEIKKKILYEGTHLDPERKHNGSSSN |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
COX20 family
|
|||||
| Subcellular location |
Mitochondrion inner membrane
|
|||||
| Function |
Essential for the assembly of the mitochondrial respiratory chain complex IV (CIV), also known as cytochrome c oxidase. Acts as a chaperone in the early steps of cytochrome c oxidase subunit II (MT-CO2/COX2) maturation, stabilizing the newly synthesized protein and presenting it to metallochaperones SCO1/2 which in turn facilitates the incorporation of the mature MT-CO2/COX2 into the assembling CIV holoenzyme.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| DEL | Deletion: p.C77Ter | . | |||
| DOTC24510 | SNV: p.E109Q | . | |||
| HEP3B217 | SNV: p.R85I | . | |||
| NUGC3 | SNV: p.R89I | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
CHEMBL5175495 Probe Info |
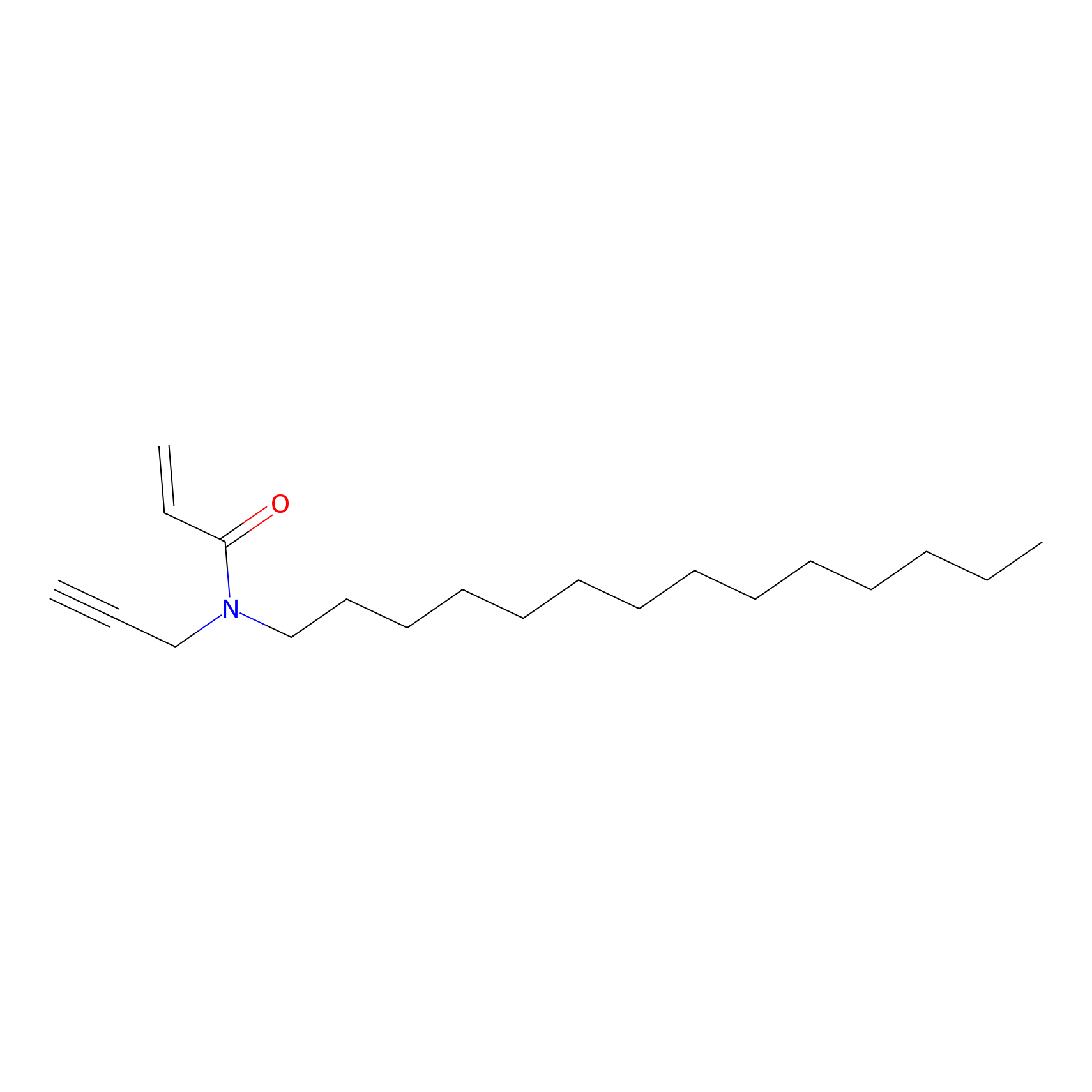 |
7.20 | LDD0196 | [1] | |
|
N1 Probe Info |
 |
100.00 | LDD0242 | [2] | |
|
STPyne Probe Info |
 |
K17(5.88); K83(0.38); K98(10.00) | LDD0277 | [3] | |
|
DBIA Probe Info |
 |
C41(1.97) | LDD3311 | [4] | |
|
JZ128-DTB Probe Info |
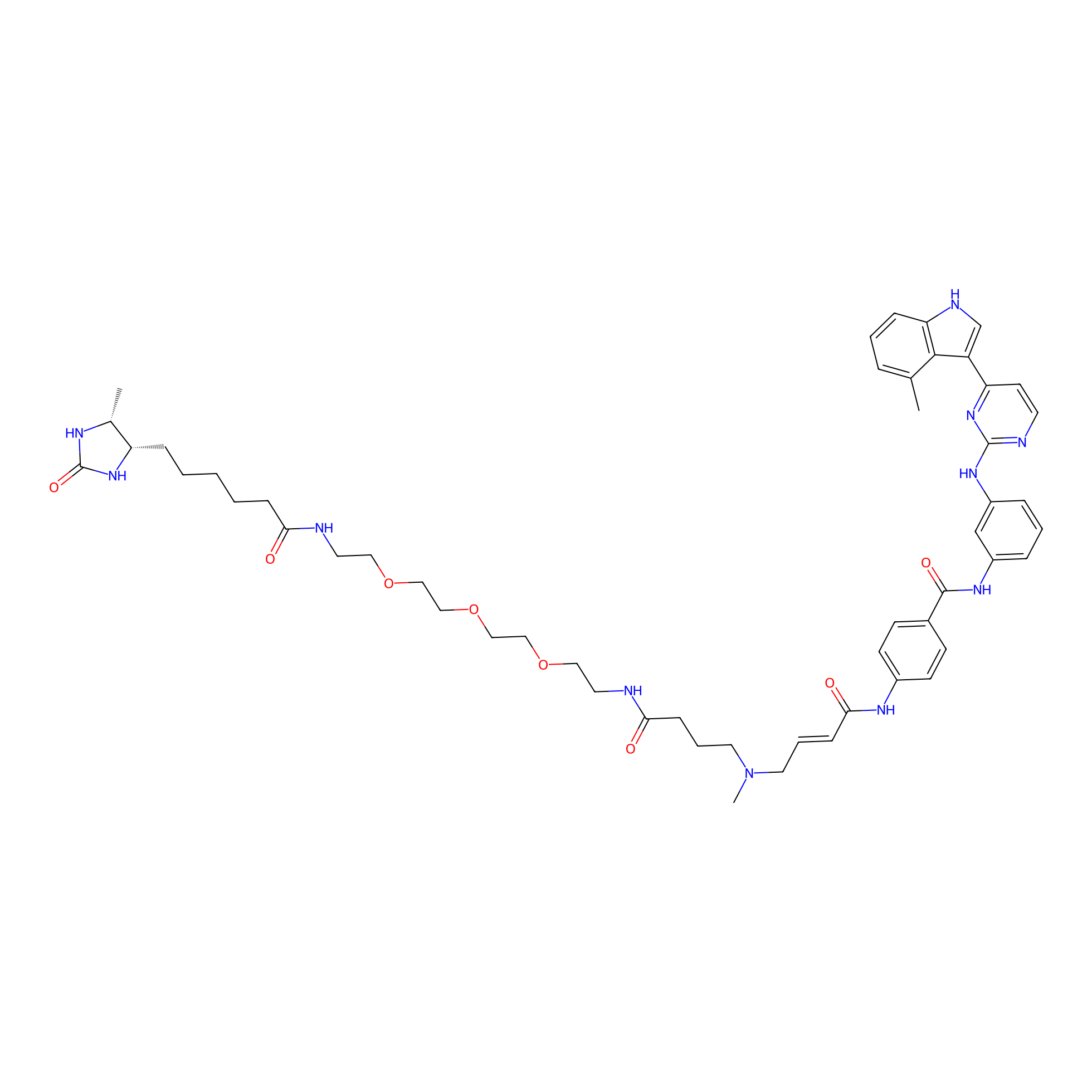 |
N.A. | LDD0462 | [5] | |
|
IPM Probe Info |
 |
C29(4.11) | LDD1701 | [6] | |
|
m-APA Probe Info |
 |
N.A. | LDD2231 | [7] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0162 | [8] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [9] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [10] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [10] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [11] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [12] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [13] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [14] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C100 Probe Info |
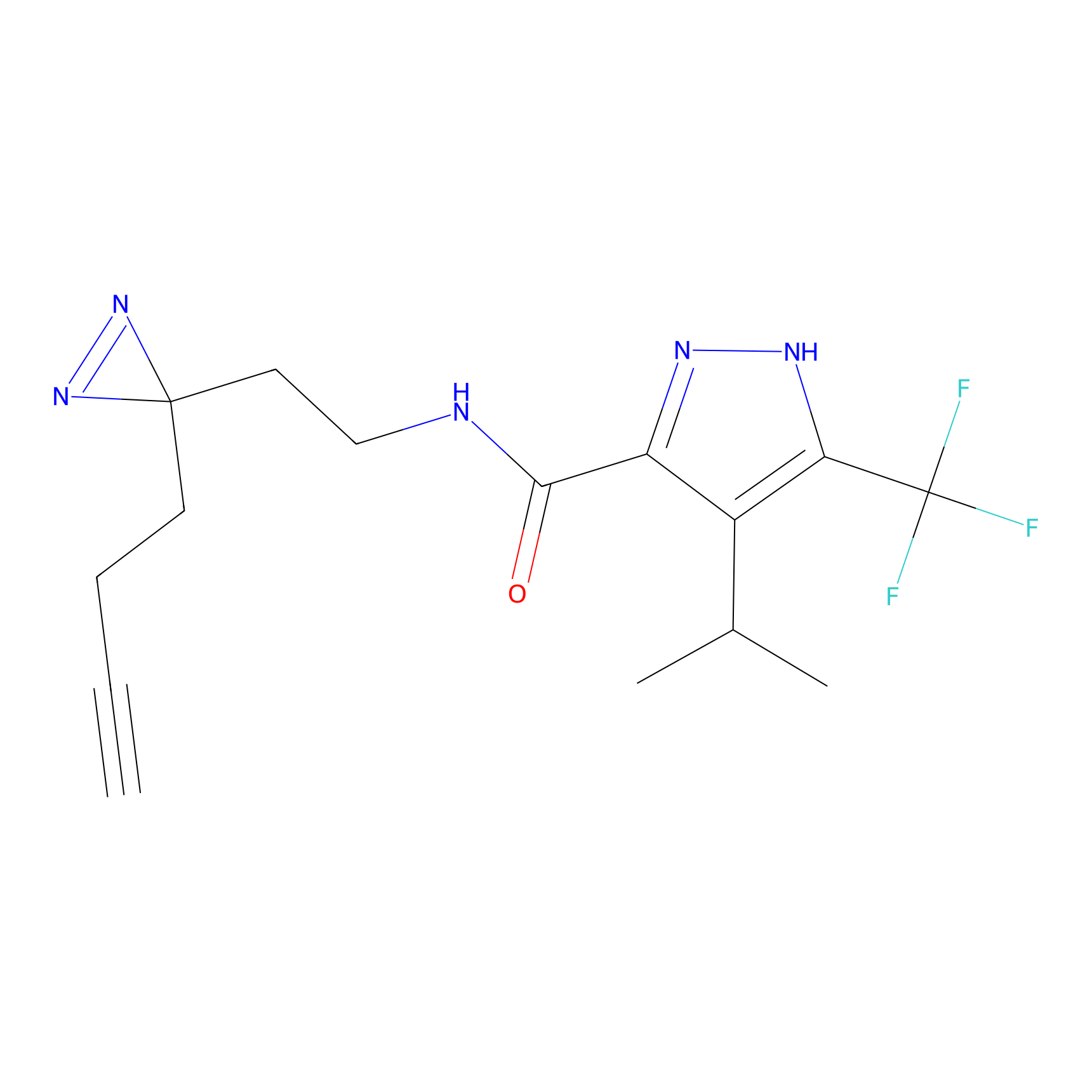 |
8.57 | LDD1789 | [15] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0215 | AC10 | HEK-293T | C29(1.01) | LDD1508 | [16] |
| LDCM0237 | AC12 | HEK-293T | C29(1.23) | LDD1510 | [16] |
| LDCM0277 | AC18 | HEK-293T | C29(1.13) | LDD1516 | [16] |
| LDCM0279 | AC2 | HEK-293T | C29(1.04) | LDD1518 | [16] |
| LDCM0280 | AC20 | HEK-293T | C29(1.15) | LDD1519 | [16] |
| LDCM0281 | AC21 | HEK-293T | C29(0.93) | LDD1520 | [16] |
| LDCM0284 | AC24 | HEK-293T | C29(1.08) | LDD1523 | [16] |
| LDCM0286 | AC26 | HEK-293T | C29(1.10) | LDD1525 | [16] |
| LDCM0288 | AC28 | HEK-293T | C29(1.03) | LDD1527 | [16] |
| LDCM0289 | AC29 | HEK-293T | C29(1.03) | LDD1528 | [16] |
| LDCM0293 | AC32 | HEK-293T | C29(0.89) | LDD1532 | [16] |
| LDCM0295 | AC34 | HEK-293T | C29(0.99) | LDD1534 | [16] |
| LDCM0297 | AC36 | HEK-293T | C29(1.45) | LDD1536 | [16] |
| LDCM0298 | AC37 | HEK-293T | C29(1.12) | LDD1537 | [16] |
| LDCM0301 | AC4 | HEK-293T | C29(1.26) | LDD1540 | [16] |
| LDCM0302 | AC40 | HEK-293T | C29(1.10) | LDD1541 | [16] |
| LDCM0304 | AC42 | HEK-293T | C29(1.03) | LDD1543 | [16] |
| LDCM0306 | AC44 | HEK-293T | C29(1.10) | LDD1545 | [16] |
| LDCM0307 | AC45 | HEK-293T | C29(1.01) | LDD1546 | [16] |
| LDCM0310 | AC48 | HEK-293T | C29(1.04) | LDD1549 | [16] |
| LDCM0312 | AC5 | HEK-293T | C29(1.03) | LDD1551 | [16] |
| LDCM0313 | AC50 | HEK-293T | C29(1.06) | LDD1552 | [16] |
| LDCM0315 | AC52 | HEK-293T | C29(1.22) | LDD1554 | [16] |
| LDCM0316 | AC53 | HEK-293T | C29(1.18) | LDD1555 | [16] |
| LDCM0319 | AC56 | HEK-293T | C29(1.08) | LDD1558 | [16] |
| LDCM0321 | AC58 | HEK-293T | C29(1.02) | LDD1560 | [16] |
| LDCM0324 | AC60 | HEK-293T | C29(0.94) | LDD1563 | [16] |
| LDCM0325 | AC61 | HEK-293T | C29(1.08) | LDD1564 | [16] |
| LDCM0328 | AC64 | HEK-293T | C29(1.15) | LDD1567 | [16] |
| LDCM0345 | AC8 | HEK-293T | C29(1.02) | LDD1569 | [16] |
| LDCM0248 | AKOS034007472 | HEK-293T | C29(0.97) | LDD1511 | [16] |
| LDCM0275 | AKOS034007705 | HEK-293T | C29(1.05) | LDD1514 | [16] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [12] |
| LDCM0372 | CL103 | HEK-293T | C29(1.15) | LDD1576 | [16] |
| LDCM0376 | CL107 | HEK-293T | C29(1.24) | LDD1580 | [16] |
| LDCM0381 | CL111 | HEK-293T | C29(1.07) | LDD1585 | [16] |
| LDCM0385 | CL115 | HEK-293T | C29(1.05) | LDD1589 | [16] |
| LDCM0389 | CL119 | HEK-293T | C29(1.18) | LDD1593 | [16] |
| LDCM0390 | CL12 | HEK-293T | C29(1.14) | LDD1594 | [16] |
| LDCM0394 | CL123 | HEK-293T | C29(0.97) | LDD1598 | [16] |
| LDCM0398 | CL127 | HEK-293T | C29(1.15) | LDD1602 | [16] |
| LDCM0402 | CL15 | HEK-293T | C29(0.97) | LDD1606 | [16] |
| LDCM0405 | CL18 | HEK-293T | C29(1.27) | LDD1609 | [16] |
| LDCM0408 | CL20 | HEK-293T | C29(1.05) | LDD1612 | [16] |
| LDCM0409 | CL21 | HEK-293T | C29(1.13) | LDD1613 | [16] |
| LDCM0412 | CL24 | HEK-293T | C29(1.23) | LDD1616 | [16] |
| LDCM0415 | CL27 | HEK-293T | C29(0.97) | LDD1619 | [16] |
| LDCM0418 | CL3 | HEK-293T | C29(0.95) | LDD1622 | [16] |
| LDCM0419 | CL30 | HEK-293T | C29(1.14) | LDD1623 | [16] |
| LDCM0421 | CL32 | HEK-293T | C29(1.23) | LDD1625 | [16] |
| LDCM0422 | CL33 | HEK-293T | C29(0.78) | LDD1626 | [16] |
| LDCM0425 | CL36 | HEK-293T | C29(1.16) | LDD1629 | [16] |
| LDCM0428 | CL39 | HEK-293T | C29(1.06) | LDD1632 | [16] |
| LDCM0432 | CL42 | HEK-293T | C29(1.11) | LDD1636 | [16] |
| LDCM0434 | CL44 | HEK-293T | C29(1.03) | LDD1638 | [16] |
| LDCM0435 | CL45 | HEK-293T | C29(1.03) | LDD1639 | [16] |
| LDCM0438 | CL48 | HEK-293T | C29(1.17) | LDD1642 | [16] |
| LDCM0445 | CL54 | HEK-293T | C29(1.13) | LDD1648 | [16] |
| LDCM0447 | CL56 | HEK-293T | C29(1.02) | LDD1650 | [16] |
| LDCM0448 | CL57 | HEK-293T | C29(0.98) | LDD1651 | [16] |
| LDCM0451 | CL6 | HEK-293T | C29(1.02) | LDD1654 | [16] |
| LDCM0452 | CL60 | HEK-293T | C29(1.11) | LDD1655 | [16] |
| LDCM0455 | CL63 | HEK-293T | C29(1.12) | LDD1658 | [16] |
| LDCM0458 | CL66 | HEK-293T | C29(1.02) | LDD1661 | [16] |
| LDCM0460 | CL68 | HEK-293T | C29(1.20) | LDD1663 | [16] |
| LDCM0461 | CL69 | HEK-293T | C29(0.97) | LDD1664 | [16] |
| LDCM0465 | CL72 | HEK-293T | C29(1.06) | LDD1668 | [16] |
| LDCM0471 | CL78 | HEK-293T | C29(0.99) | LDD1674 | [16] |
| LDCM0473 | CL8 | HEK-293T | C29(1.24) | LDD1676 | [16] |
| LDCM0474 | CL80 | HEK-293T | C29(1.49) | LDD1677 | [16] |
| LDCM0475 | CL81 | HEK-293T | C29(1.15) | LDD1678 | [16] |
| LDCM0478 | CL84 | HEK-293T | C29(1.02) | LDD1681 | [16] |
| LDCM0481 | CL87 | HEK-293T | C29(1.06) | LDD1684 | [16] |
| LDCM0484 | CL9 | HEK-293T | C29(1.27) | LDD1687 | [16] |
| LDCM0485 | CL90 | HEK-293T | C29(0.87) | LDD1688 | [16] |
| LDCM0487 | CL92 | HEK-293T | C29(1.03) | LDD1690 | [16] |
| LDCM0488 | CL93 | HEK-293T | C29(1.01) | LDD1691 | [16] |
| LDCM0491 | CL96 | HEK-293T | C29(1.14) | LDD1694 | [16] |
| LDCM0494 | CL99 | HEK-293T | C29(0.86) | LDD1697 | [16] |
| LDCM0495 | E2913 | HEK-293T | C29(1.12) | LDD1698 | [16] |
| LDCM0625 | F8 | Ramos | C29(0.57) | LDD2187 | [17] |
| LDCM0572 | Fragment10 | Ramos | C29(1.17) | LDD2189 | [17] |
| LDCM0573 | Fragment11 | Ramos | C29(0.02) | LDD2190 | [17] |
| LDCM0574 | Fragment12 | Ramos | C29(2.00) | LDD2191 | [17] |
| LDCM0575 | Fragment13 | Ramos | C29(0.92) | LDD2192 | [17] |
| LDCM0576 | Fragment14 | Ramos | C29(2.50) | LDD2193 | [17] |
| LDCM0579 | Fragment20 | Ramos | C29(2.12) | LDD2194 | [17] |
| LDCM0580 | Fragment21 | Ramos | C29(1.07) | LDD2195 | [17] |
| LDCM0582 | Fragment23 | Ramos | C29(1.66) | LDD2196 | [17] |
| LDCM0578 | Fragment27 | Ramos | C29(0.84) | LDD2197 | [17] |
| LDCM0586 | Fragment28 | Ramos | C29(0.55) | LDD2198 | [17] |
| LDCM0588 | Fragment30 | Ramos | C29(0.85) | LDD2199 | [17] |
| LDCM0589 | Fragment31 | Ramos | C29(1.29) | LDD2200 | [17] |
| LDCM0590 | Fragment32 | Ramos | C29(1.30) | LDD2201 | [17] |
| LDCM0468 | Fragment33 | HEK-293T | C29(1.13) | LDD1671 | [16] |
| LDCM0596 | Fragment38 | Ramos | C29(1.13) | LDD2203 | [17] |
| LDCM0566 | Fragment4 | Ramos | C29(1.49) | LDD2184 | [17] |
| LDCM0610 | Fragment52 | Ramos | C29(0.82) | LDD2204 | [17] |
| LDCM0614 | Fragment56 | Ramos | C29(0.81) | LDD2205 | [17] |
| LDCM0569 | Fragment7 | Ramos | C29(1.15) | LDD2186 | [17] |
| LDCM0571 | Fragment9 | Ramos | C29(2.63) | LDD2188 | [17] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [12] |
| LDCM0179 | JZ128 | PC-3 | N.A. | LDD0462 | [5] |
| LDCM0022 | KB02 | Ramos | C29(1.09) | LDD2182 | [17] |
| LDCM0023 | KB03 | MDA-MB-231 | C29(4.11) | LDD1701 | [6] |
| LDCM0024 | KB05 | G361 | C41(1.97) | LDD3311 | [4] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [12] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Cyclic AMP-responsive element-binding protein 3-like protein 1 (CREB3L1) | BZIP family | Q96BA8 | |||
Other
References
