Details of the Target
General Information of Target
| Target ID | LDTP07042 | |||||
|---|---|---|---|---|---|---|
| Target Name | Protein Smaug homolog 2 (SAMD4B) | |||||
| Gene Name | SAMD4B | |||||
| Gene ID | 55095 | |||||
| Synonyms |
SMAUG2; Protein Smaug homolog 2; Smaug 2; hSmaug2; Sterile alpha motif domain-containing protein 4B; SAM domain-containing protein 4B |
|||||
| 3D Structure | ||||||
| Sequence |
MMFRDQVGILAGWFKGWNECEQTVALLSLLKRVTRTQARFLQLCLEHSLADCNDIHLLES
EANSAAIVSQWQQESKEKVVSLLLSHLPLLQPGNTEAKSEYMRLLQKVLAYSIESNAFIE ESRQLLSYALIHPATTLEDRNALALWLSHLEERLASGFRSRPEPSYHSRQGSDEWGGPAE LGPGEAGPGWQDKPPRENGHVPFHPSSSVPPAINSIGSNANTGLPCQIHPSPLKRSMSLI PTSPQVPGEWPSPEELGARAAFTTPDHAPLSPQSSVASSGSEQTEEQGSSRNTFQEDGSG MKDVPSWLKSLRLHKYAALFSQMSYEEMMTLTEQHLESQNVTKGARHKIALSIQKLRERQ SVLKSLEKDVLEGGNLRNALQELQQIIITPIKAYSVLQATVAAATTTPTAKDGAPGEPPL PGAEPPLAHPGTDKGTEAKDPPAVENYPPPPAPAPTDGSEPAPAPVADGDIPSQFTRVMG KVCTQLLVSRPDEENITSYLQLIEKCLTHEAFTETQKKRLLSWKQQVLKLLRTFPRKAAL EMQNYRQQKGWAFGSNSLPIAGSVGMGVARRTQRQFPMPPRALPPGRMGLLSPSGIGGVS PRHALTSPSLGGQGRQNLWFANPGGSNSMPSQSRSSVQRTHSLPVHSSPQAILMFPPDCP VPGPDLEINPTLESLCLSMTEHALGDGTDKTSTI |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
SMAUG family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function | Has transcriptional repressor activity. Overexpression inhibits the transcriptional activities of AP-1, p53/TP53 and CDKN1A. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
STPyne Probe Info |
 |
K524(8.28) | LDD0277 | [2] | |
|
DBIA Probe Info |
 |
C20(3.77) | LDD3365 | [3] | |
|
AHL-Pu-1 Probe Info |
 |
C20(2.38) | LDD0171 | [4] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [5] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0162 | [6] | |
|
Lodoacetamide azide Probe Info |
 |
C483(0.00); C20(0.00) | LDD0037 | [5] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [7] | |
|
IPM Probe Info |
 |
C226(0.00); C20(0.00) | LDD2156 | [8] | |
|
AOyne Probe Info |
 |
14.00 | LDD0443 | [9] | |
|
NAIA_5 Probe Info |
 |
C506(0.00); C20(0.00); C483(0.00) | LDD2223 | [7] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C266 Probe Info |
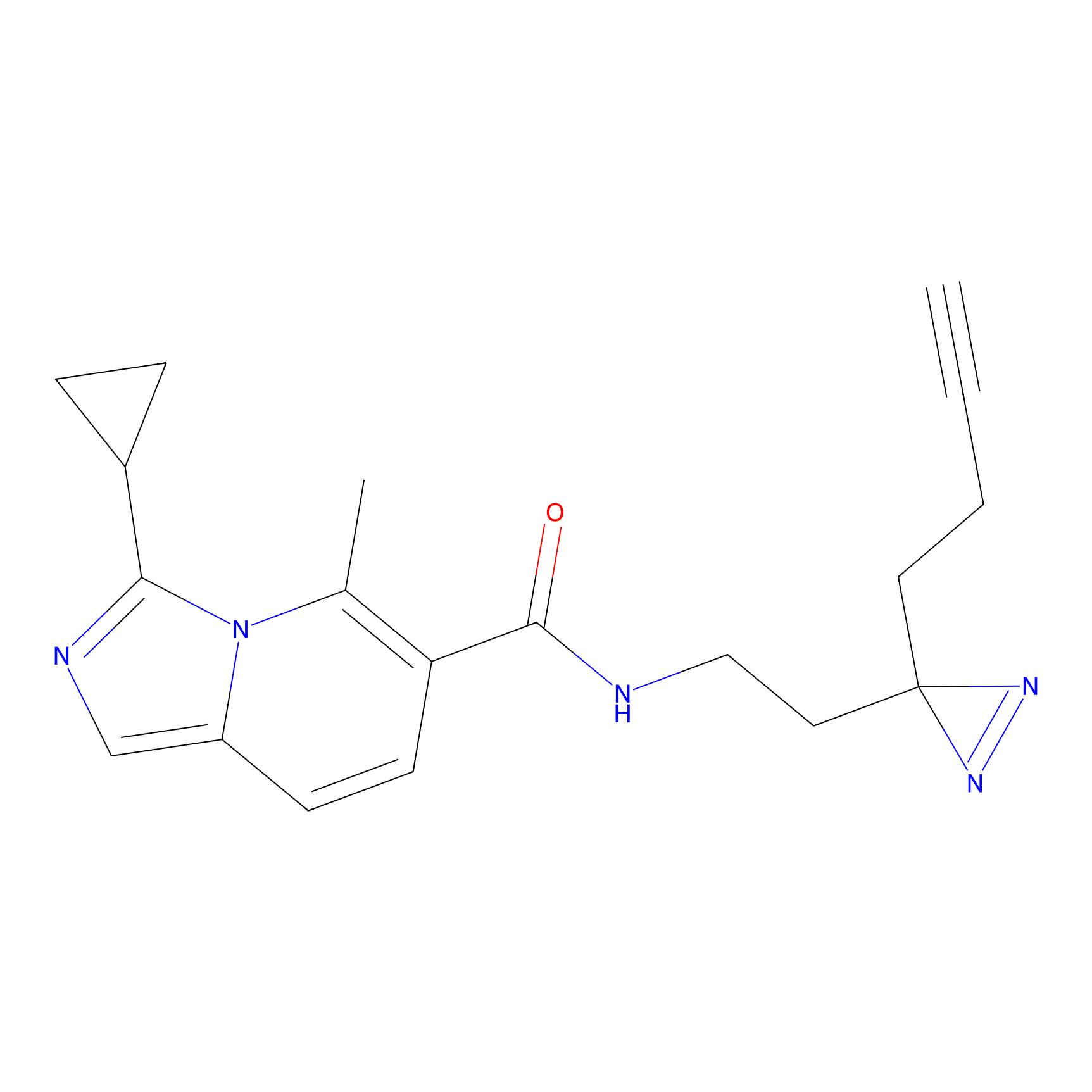 |
11.47 | LDD1937 | [10] | |
|
C362 Probe Info |
 |
28.84 | LDD2023 | [10] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0026 | 4SU-RNA+native RNA | DM93 | C20(2.38) | LDD0171 | [4] |
| LDCM0214 | AC1 | PaTu 8988t | C506(0.24) | LDD1093 | [11] |
| LDCM0215 | AC10 | PaTu 8988t | C506(0.72) | LDD1094 | [11] |
| LDCM0226 | AC11 | PaTu 8988t | C506(0.44) | LDD1105 | [11] |
| LDCM0237 | AC12 | PaTu 8988t | C506(0.47) | LDD1116 | [11] |
| LDCM0259 | AC14 | PaTu 8988t | C506(0.49) | LDD1138 | [11] |
| LDCM0270 | AC15 | PaTu 8988t | C506(0.64) | LDD1149 | [11] |
| LDCM0279 | AC2 | PaTu 8988t | C506(0.38) | LDD1158 | [11] |
| LDCM0290 | AC3 | PaTu 8988t | C506(0.26) | LDD1169 | [11] |
| LDCM0301 | AC4 | PaTu 8988t | C506(0.31) | LDD1180 | [11] |
| LDCM0312 | AC5 | PaTu 8988t | C506(0.26) | LDD1191 | [11] |
| LDCM0323 | AC6 | PaTu 8988t | C506(0.72) | LDD1202 | [11] |
| LDCM0334 | AC7 | PaTu 8988t | C506(0.85) | LDD1213 | [11] |
| LDCM0345 | AC8 | PaTu 8988t | C506(1.02) | LDD1224 | [11] |
| LDCM0248 | AKOS034007472 | PaTu 8988t | C506(0.47) | LDD1127 | [11] |
| LDCM0356 | AKOS034007680 | PaTu 8988t | C506(0.90) | LDD1235 | [11] |
| LDCM0275 | AKOS034007705 | PaTu 8988t | C506(0.81) | LDD1154 | [11] |
| LDCM0156 | Aniline | NCI-H1299 | 11.57 | LDD0403 | [1] |
| LDCM0632 | CL-Sc | Hep-G2 | C20(0.18) | LDD2227 | [7] |
| LDCM0367 | CL1 | PaTu 8988t | C506(1.06) | LDD1246 | [11] |
| LDCM0368 | CL10 | PaTu 8988t | C506(0.60) | LDD1247 | [11] |
| LDCM0369 | CL100 | PaTu 8988t | C506(0.33) | LDD1248 | [11] |
| LDCM0370 | CL101 | PaTu 8988t | C506(0.79) | LDD1249 | [11] |
| LDCM0371 | CL102 | PaTu 8988t | C506(0.81) | LDD1250 | [11] |
| LDCM0372 | CL103 | PaTu 8988t | C506(0.44) | LDD1251 | [11] |
| LDCM0373 | CL104 | PaTu 8988t | C506(0.54) | LDD1252 | [11] |
| LDCM0375 | CL106 | HEK-293T | C52(0.97) | LDD1579 | [12] |
| LDCM0377 | CL108 | HEK-293T | C52(0.89) | LDD1581 | [12] |
| LDCM0379 | CL11 | PaTu 8988t | C506(0.96) | LDD1258 | [11] |
| LDCM0380 | CL110 | HEK-293T | C52(1.32) | LDD1584 | [12] |
| LDCM0382 | CL112 | HEK-293T | C52(0.97) | LDD1586 | [12] |
| LDCM0384 | CL114 | HEK-293T | C52(1.14) | LDD1588 | [12] |
| LDCM0386 | CL116 | HEK-293T | C52(1.00) | LDD1590 | [12] |
| LDCM0388 | CL118 | HEK-293T | C52(1.30) | LDD1592 | [12] |
| LDCM0390 | CL12 | PaTu 8988t | C506(0.82) | LDD1269 | [11] |
| LDCM0391 | CL120 | HEK-293T | C52(1.00) | LDD1595 | [12] |
| LDCM0393 | CL122 | HEK-293T | C52(1.11) | LDD1597 | [12] |
| LDCM0395 | CL124 | HEK-293T | C52(1.07) | LDD1599 | [12] |
| LDCM0397 | CL126 | HEK-293T | C52(1.42) | LDD1601 | [12] |
| LDCM0399 | CL128 | HEK-293T | C52(0.99) | LDD1603 | [12] |
| LDCM0400 | CL13 | PaTu 8988t | C506(0.51) | LDD1279 | [11] |
| LDCM0401 | CL14 | PaTu 8988t | C506(0.51) | LDD1280 | [11] |
| LDCM0402 | CL15 | PaTu 8988t | C506(0.79) | LDD1281 | [11] |
| LDCM0403 | CL16 | HEK-293T | C52(1.06) | LDD1607 | [12] |
| LDCM0407 | CL2 | PaTu 8988t | C506(0.68) | LDD1286 | [11] |
| LDCM0414 | CL26 | HEK-293T | C52(1.24) | LDD1618 | [12] |
| LDCM0416 | CL28 | HEK-293T | C52(1.09) | LDD1620 | [12] |
| LDCM0418 | CL3 | PaTu 8988t | C506(1.11) | LDD1297 | [11] |
| LDCM0429 | CL4 | PaTu 8988t | C506(1.36) | LDD1308 | [11] |
| LDCM0430 | CL40 | HEK-293T | C52(0.92) | LDD1634 | [12] |
| LDCM0436 | CL46 | PaTu 8988t | C506(1.00) | LDD1315 | [11] |
| LDCM0437 | CL47 | PaTu 8988t | C506(0.97) | LDD1316 | [11] |
| LDCM0438 | CL48 | PaTu 8988t | C506(0.94) | LDD1317 | [11] |
| LDCM0439 | CL49 | PaTu 8988t | C506(0.97) | LDD1318 | [11] |
| LDCM0440 | CL5 | PaTu 8988t | C506(0.62) | LDD1319 | [11] |
| LDCM0441 | CL50 | PaTu 8988t | C506(0.94) | LDD1320 | [11] |
| LDCM0442 | CL51 | PaTu 8988t | C506(0.99) | LDD1321 | [11] |
| LDCM0443 | CL52 | PaTu 8988t | C506(0.88) | LDD1322 | [11] |
| LDCM0444 | CL53 | PaTu 8988t | C506(0.81) | LDD1323 | [11] |
| LDCM0445 | CL54 | PaTu 8988t | C506(1.01) | LDD1324 | [11] |
| LDCM0446 | CL55 | PaTu 8988t | C506(1.10) | LDD1325 | [11] |
| LDCM0447 | CL56 | PaTu 8988t | C506(0.25) | LDD1326 | [11] |
| LDCM0448 | CL57 | PaTu 8988t | C506(0.49) | LDD1327 | [11] |
| LDCM0449 | CL58 | PaTu 8988t | C506(0.84) | LDD1328 | [11] |
| LDCM0450 | CL59 | PaTu 8988t | C506(0.55) | LDD1329 | [11] |
| LDCM0451 | CL6 | PaTu 8988t | C506(0.58) | LDD1330 | [11] |
| LDCM0452 | CL60 | PaTu 8988t | C506(0.55) | LDD1331 | [11] |
| LDCM0454 | CL62 | HEK-293T | C52(1.04) | LDD1657 | [12] |
| LDCM0456 | CL64 | HEK-293T | C52(1.29) | LDD1659 | [12] |
| LDCM0462 | CL7 | PaTu 8988t | C506(0.57) | LDD1341 | [11] |
| LDCM0467 | CL74 | HEK-293T | C52(1.17) | LDD1670 | [12] |
| LDCM0469 | CL76 | HEK-293T | C52(1.15) | LDD1672 | [12] |
| LDCM0473 | CL8 | PaTu 8988t | C506(0.93) | LDD1352 | [11] |
| LDCM0480 | CL86 | HEK-293T | C52(1.52) | LDD1683 | [12] |
| LDCM0482 | CL88 | HEK-293T | C52(0.99) | LDD1685 | [12] |
| LDCM0484 | CL9 | PaTu 8988t | C506(0.49) | LDD1363 | [11] |
| LDCM0486 | CL91 | PaTu 8988t | C506(0.36) | LDD1365 | [11] |
| LDCM0487 | CL92 | PaTu 8988t | C506(0.33) | LDD1366 | [11] |
| LDCM0488 | CL93 | PaTu 8988t | C506(0.37) | LDD1367 | [11] |
| LDCM0489 | CL94 | PaTu 8988t | C506(0.34) | LDD1368 | [11] |
| LDCM0490 | CL95 | PaTu 8988t | C506(0.38) | LDD1369 | [11] |
| LDCM0491 | CL96 | PaTu 8988t | C506(0.40) | LDD1370 | [11] |
| LDCM0492 | CL97 | PaTu 8988t | C506(0.42) | LDD1371 | [11] |
| LDCM0493 | CL98 | PaTu 8988t | C506(0.32) | LDD1372 | [11] |
| LDCM0494 | CL99 | PaTu 8988t | C506(0.35) | LDD1373 | [11] |
| LDCM0427 | Fragment51 | HEK-293T | C52(1.28) | LDD1631 | [12] |
| LDCM0022 | KB02 | A2780 | C20(2.66) | LDD2254 | [3] |
| LDCM0023 | KB03 | A2780 | C20(4.35) | LDD2671 | [3] |
| LDCM0024 | KB05 | OVCAR-3 | C20(3.77) | LDD3365 | [3] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Inactive ubiquitin carboxyl-terminal hydrolase 53 (USP53) | Peptidase C19 family | Q70EK8 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| 14-3-3 protein zeta/delta (YWHAZ) | 14-3-3 family | P63104 | |||
| Volume-regulated anion channel subunit LRRC8E (LRRC8E) | LRRC8 family | Q6NSJ5 | |||
Other
References
