Details of the Target
General Information of Target
| Target ID | LDTP06350 | |||||
|---|---|---|---|---|---|---|
| Target Name | RNA-binding protein with serine-rich domain 1 (RNPS1) | |||||
| Gene Name | RNPS1 | |||||
| Gene ID | 10921 | |||||
| Synonyms |
LDC2; RNA-binding protein with serine-rich domain 1; SR-related protein LDC2 |
|||||
| 3D Structure | ||||||
| Sequence |
MDLSGVKKKSLLGVKENNKKSSTRAPSPTKRKDRSDEKSKDRSKDKGATKESSEKDRGRD
KTRKRRSASSGSSSTRSRSSSTSSSGSSTSTGSSSGSSSSSASSRSGSSSTSRSSSSSSS SGSPSPSRRRHDNRRRSRSKSKPPKRDEKERKRRSPSPKPTKVHIGRLTRNVTKDHIMEI FSTYGKIKMIDMPVERMHPHLSKGYAYVEFENPDEAEKALKHMDGGQIDGQEITATAVLA PWPRPPPRRFSPPRRMLPPPPMWRRSPPRMRRRSRSPRRRSPVRRRSRSPGRRRHRSRSS SNSSR |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Splicing factor SR family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Part of pre- and post-splicing multiprotein mRNP complexes. Auxiliary component of the splicing-dependent multiprotein exon junction complex (EJC) deposited at splice junction on mRNAs. The EJC is a dynamic structure consisting of core proteins and several peripheral nuclear and cytoplasmic associated factors that join the complex only transiently either during EJC assembly or during subsequent mRNA metabolism. Component of the ASAP and PSAP complexes which bind RNA in a sequence-independent manner and are proposed to be recruited to the EJC prior to or during the splicing process and to regulate specific excision of introns in specific transcription subsets. The ASAP complex can inhibit RNA processing during in vitro splicing reactions. The ASAP complex promotes apoptosis and is disassembled after induction of apoptosis. Enhances the formation of the ATP-dependent A complex of the spliceosome. Involved in both constitutive splicing and, in association with SRP54 and TRA2B/SFRS10, in distinctive modulation of alternative splicing in a substrate-dependent manner. Involved in the splicing modulation of BCL2L1/Bcl-X (and probably other apoptotic genes); specifically inhibits formation of proapoptotic isoforms such as Bcl-X(S); the activity is different from the established EJC assembly and function. Participates in mRNA 3'-end cleavage. Involved in UPF2-dependent nonsense-mediated decay (NMD) of mRNAs containing premature stop codons. Also mediates increase of mRNA abundance and translational efficiency. Binds spliced mRNA 20-25 nt upstream of exon-exon junctions.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
P1 Probe Info |
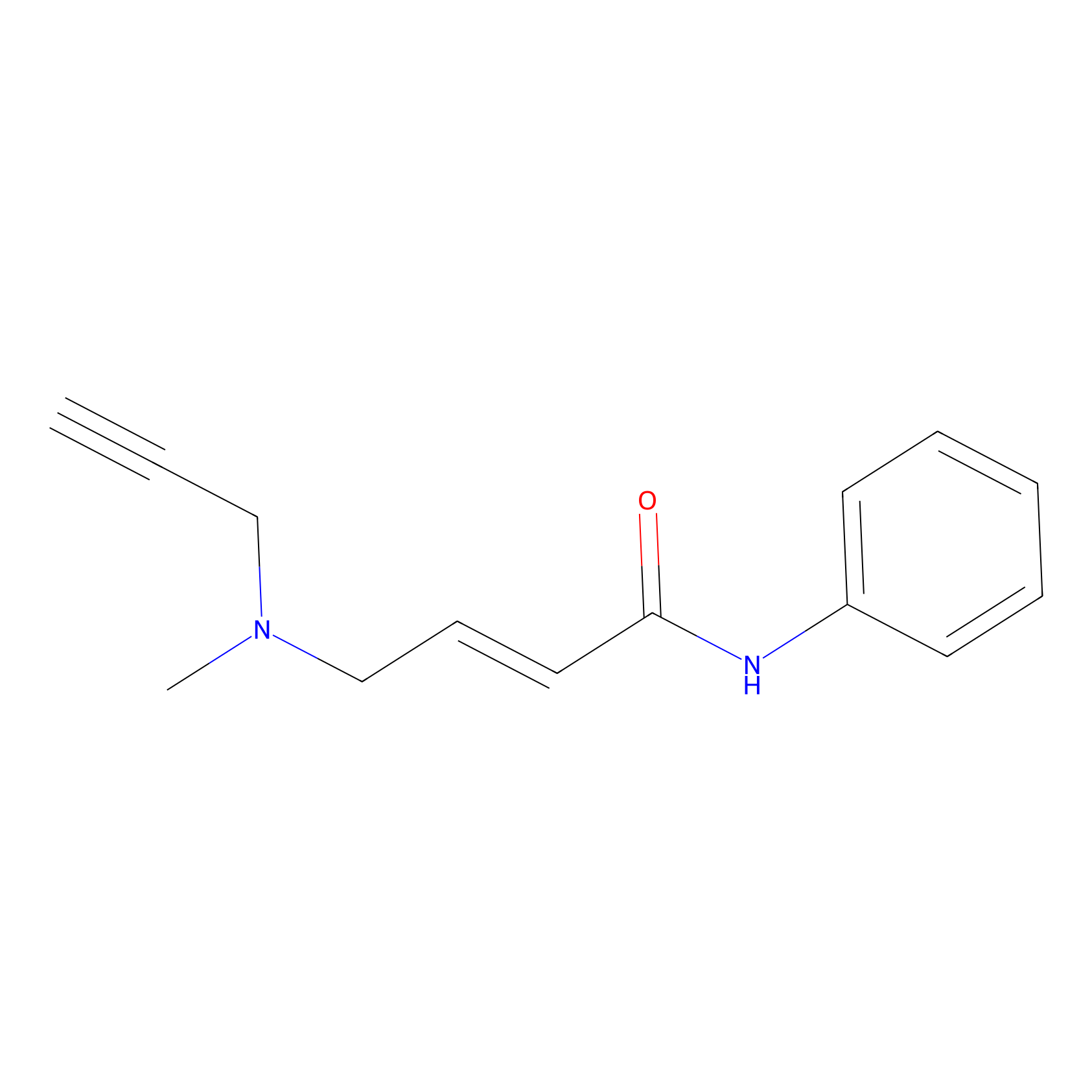 |
2.74 | LDD0448 | [1] | |
|
P2 Probe Info |
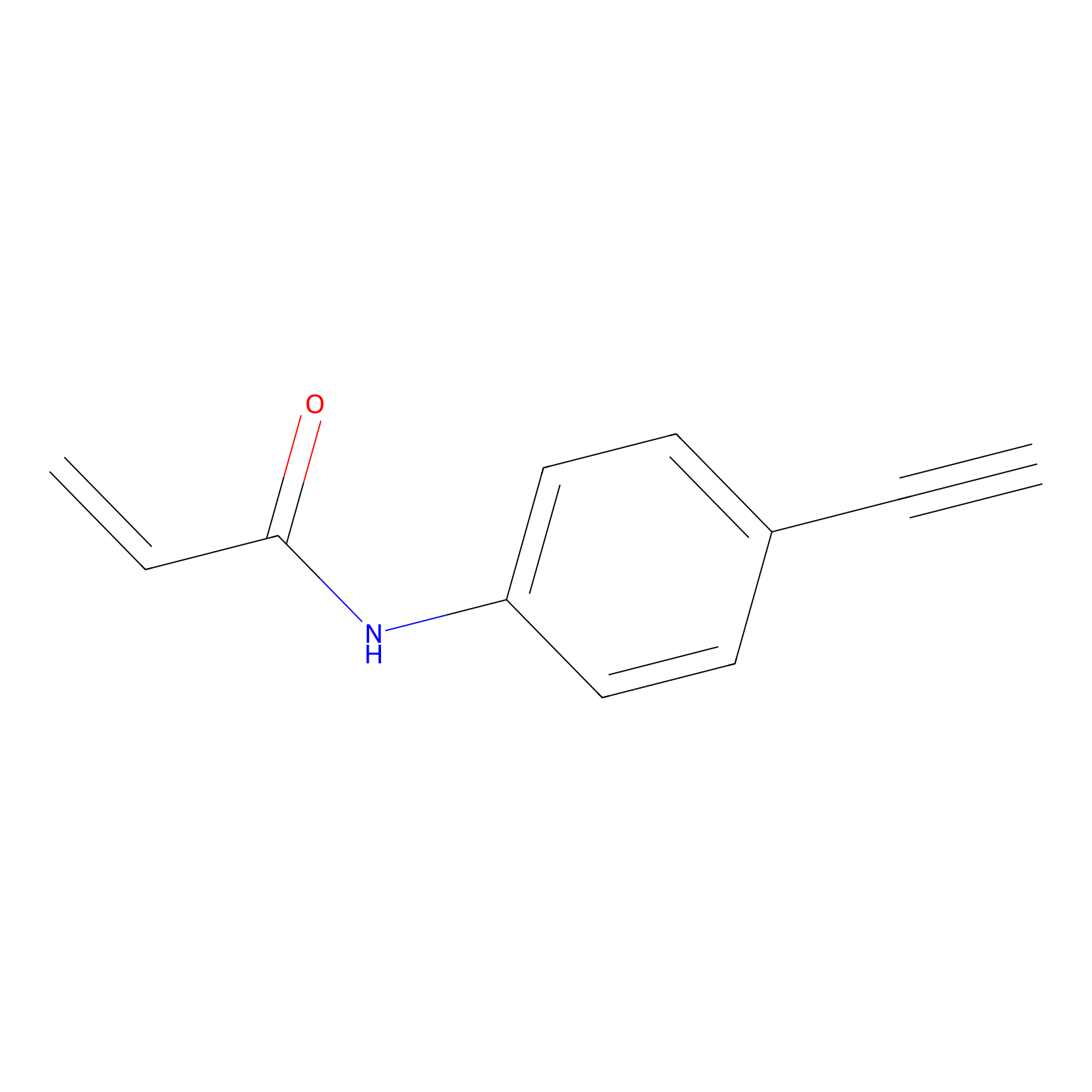 |
2.25 | LDD0449 | [1] | |
|
P3 Probe Info |
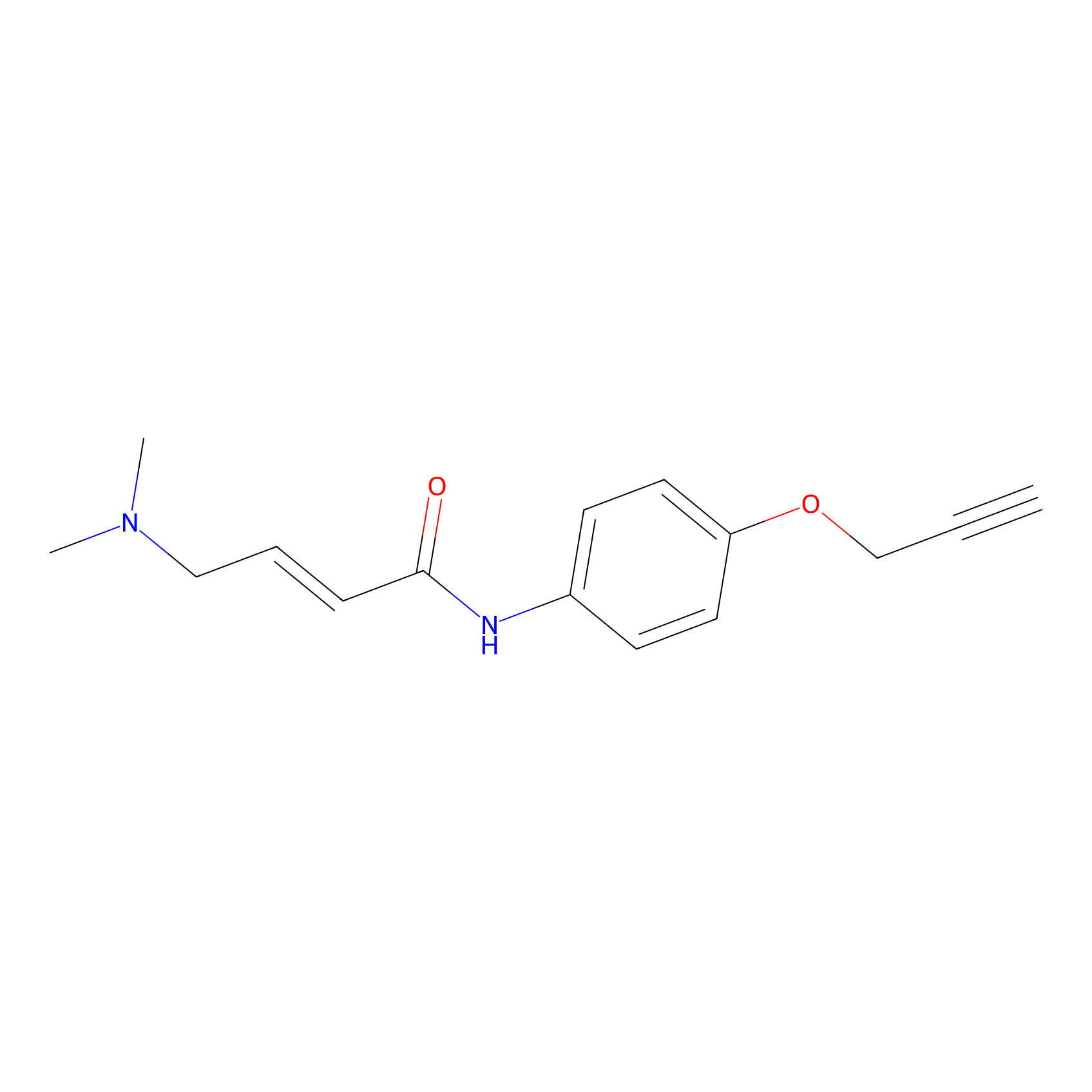 |
2.30 | LDD0450 | [1] | |
|
P8 Probe Info |
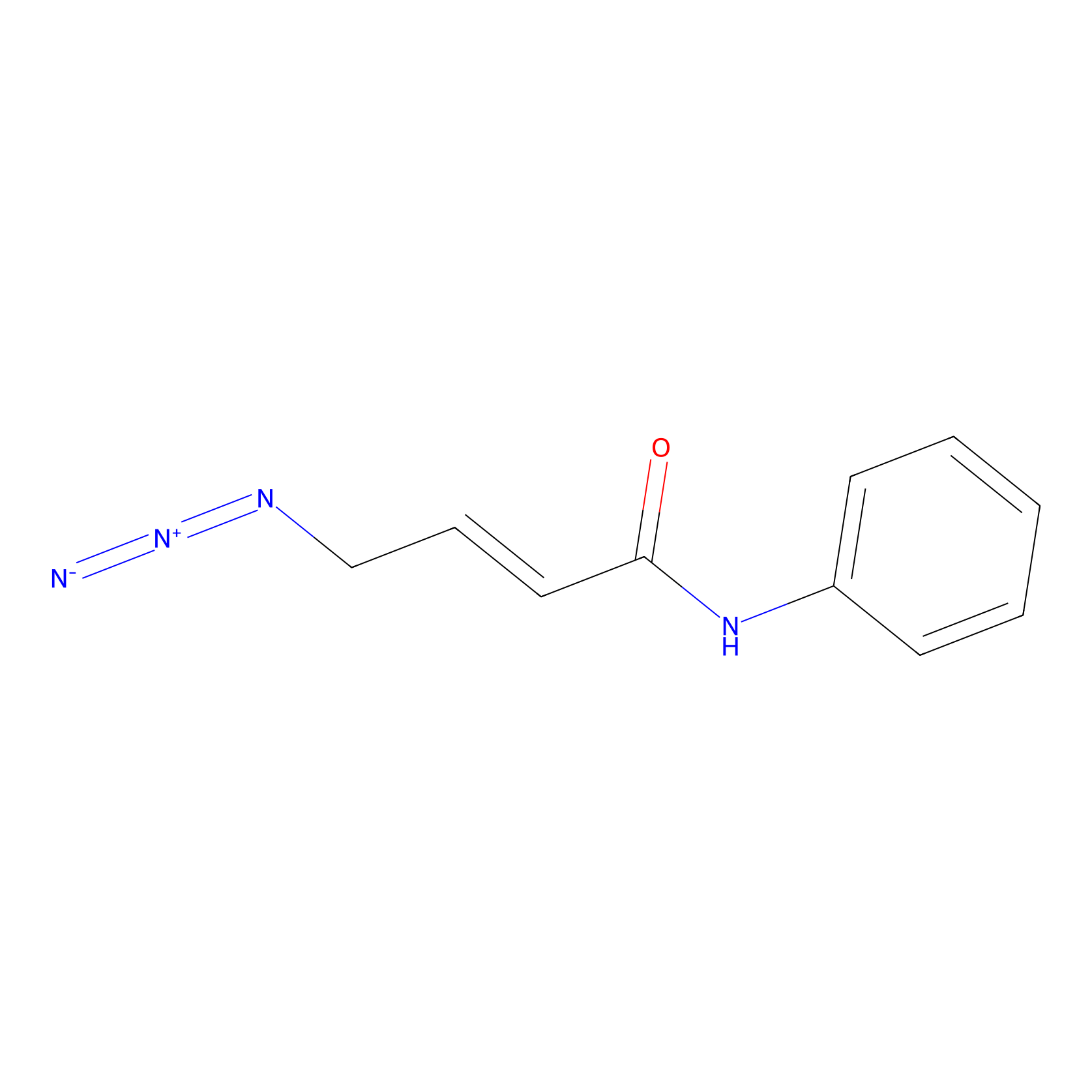 |
3.33 | LDD0451 | [1] | |
|
C-Sul Probe Info |
 |
6.85 | LDD0066 | [2] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [3] | |
|
HHS-475 Probe Info |
 |
Y207(1.74) | LDD0264 | [4] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0221 | [5] | |
|
ATP probe Probe Info |
 |
K218(0.00); K221(0.00); K203(0.00); K174(0.00) | LDD0199 | [6] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0358 | [7] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [8] | |
|
SF Probe Info |
 |
Y207(0.00); Y205(0.00) | LDD0028 | [9] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [8] | |
|
1c-yne Probe Info |
 |
K188(0.00); K186(0.00); K174(0.00) | LDD0228 | [7] | |
|
HHS-465 Probe Info |
 |
N.A. | LDD2240 | [10] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C364 Probe Info |
 |
15.14 | LDD2025 | [11] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [5] |
| LDCM0116 | HHS-0101 | DM93 | Y207(1.74) | LDD0264 | [4] |
| LDCM0117 | HHS-0201 | DM93 | Y207(0.85) | LDD0265 | [4] |
| LDCM0118 | HHS-0301 | DM93 | Y207(0.93) | LDD0266 | [4] |
| LDCM0119 | HHS-0401 | DM93 | Y207(1.11) | LDD0267 | [4] |
| LDCM0120 | HHS-0701 | DM93 | Y207(0.37) | LDD0268 | [4] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [5] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [5] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Pinin (PNN) | Pinin family | Q9H307 | |||
Transcription factor
Other
References
