Details of the Target
General Information of Target
| Target ID | LDTP05418 | |||||
|---|---|---|---|---|---|---|
| Target Name | UBX domain-containing protein 1 (UBXN1) | |||||
| Gene Name | UBXN1 | |||||
| Gene ID | 51035 | |||||
| Synonyms |
SAKS1; UBX domain-containing protein 1; SAPK substrate protein 1; UBA/UBX 33.3 kDa protein |
|||||
| 3D Structure | ||||||
| Sequence |
MAELTALESLIEMGFPRGRAEKALALTGNQGIEAAMDWLMEHEDDPDVDEPLETPLGHIL
GREPTSSEQGGLEGSGSAAGEGKPALSEEERQEQTKRMLELVAQKQREREEREEREALER ERQRRRQGQELSAARQRLQEDEMRRAAEERRREKAEELAARQRVREKIERDKAERAKKYG GSVGSQPPPVAPEPGPVPSSPSQEPPTKREYDQCRIQVRLPDGTSLTQTFRAREQLAAVR LYVELHRGEELGGGQDPVQLLSGFPRRAFSEADMERPLQELGLVPSAVLIVAKKCPS |
|||||
| Target Bioclass |
Other
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Ubiquitin-binding protein that plays a role in the modulation of innate immune response. Blocks both the RIG-I-like receptors (RLR) and NF-kappa-B pathways. Following viral infection, UBXN1 is induced and recruited to the RLR component MAVS. In turn, interferes with MAVS oligomerization, and disrupts the MAVS/TRAF3/TRAF6 signalosome. This function probably serves as a brake to prevent excessive RLR signaling. Interferes with the TNFalpha-triggered NF-kappa-B pathway by interacting with cellular inhibitors of apoptosis proteins (cIAPs) and thereby inhibiting their recruitment to TNFR1. Prevents also the activation of NF-kappa-B by associating with CUL1 and thus inhibiting NF-kappa-B inhibitor alpha/NFKBIA degradation that remains bound to NF-kappa-B. Interacts with the BRCA1-BARD1 heterodimer and regulates its activity. Specifically binds 'Lys-6'-linked polyubiquitin chains. Interaction with autoubiquitinated BRCA1 leads to the inhibition of the E3 ubiquitin-protein ligase activity of the BRCA1-BARD1 heterodimer. Component of a complex required to couple deglycosylation and proteasome-mediated degradation of misfolded proteins in the endoplasmic reticulum that are retrotranslocated in the cytosol.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
ONAyne Probe Info |
 |
K154(6.25) | LDD0274 | [2] | |
|
STPyne Probe Info |
 |
K105(10.00) | LDD0277 | [2] | |
|
DBIA Probe Info |
 |
C214(1.48) | LDD3410 | [3] | |
|
BTD Probe Info |
 |
C214(1.00) | LDD2107 | [4] | |
|
DA-P3 Probe Info |
 |
6.27 | LDD0180 | [5] | |
|
HHS-475 Probe Info |
 |
Y179(13.74) | LDD0268 | [6] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [7] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0199 | [8] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0162 | [9] | |
|
IPIAA_L Probe Info |
 |
N.A. | LDD0031 | [10] | |
|
IPM Probe Info |
 |
N.A. | LDD0147 | [11] | |
|
SF Probe Info |
 |
Y242(0.00); Y179(0.00) | LDD0028 | [12] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [11] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [13] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [14] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [14] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [14] | |
|
AOyne Probe Info |
 |
13.10 | LDD0443 | [15] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C201 Probe Info |
 |
25.63 | LDD1877 | [16] | |
|
C244 Probe Info |
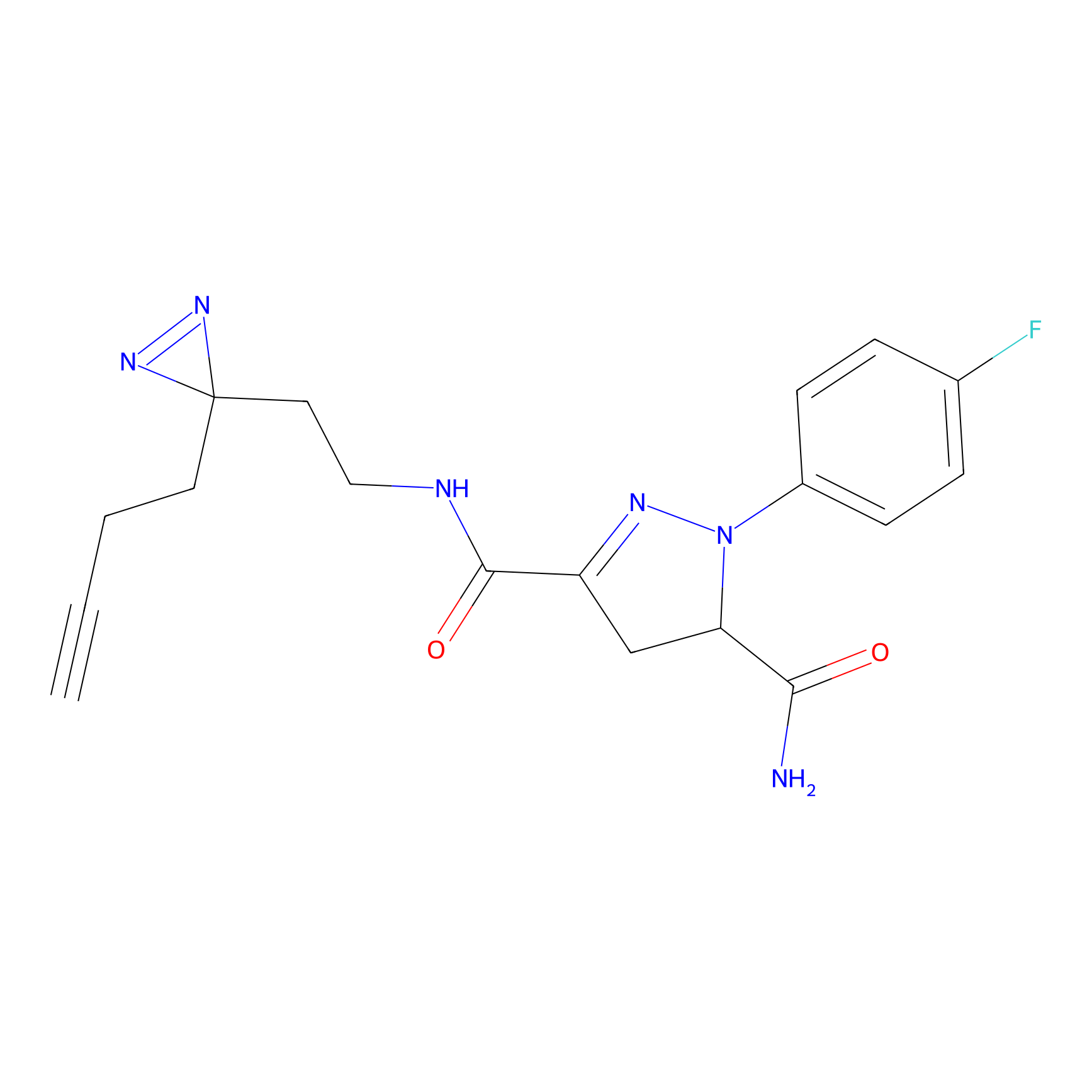 |
13.83 | LDD1917 | [16] | |
|
C284 Probe Info |
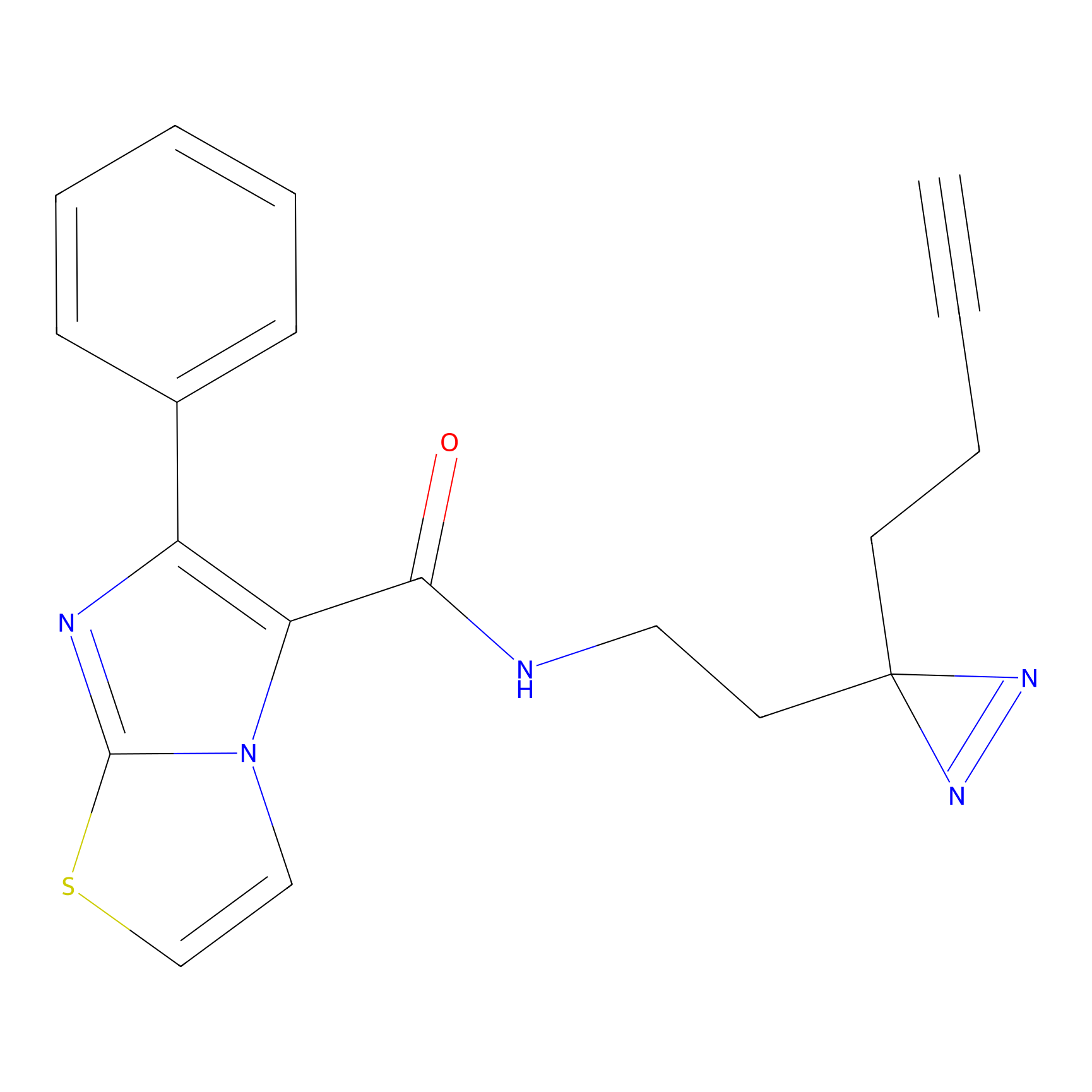 |
20.97 | LDD1954 | [16] | |
|
FFF probe11 Probe Info |
 |
20.00 | LDD0471 | [17] | |
|
FFF probe2 Probe Info |
 |
20.00 | LDD0463 | [17] | |
|
FFF probe3 Probe Info |
 |
20.00 | LDD0464 | [17] | |
|
DA-2 Probe Info |
 |
N.A. | LDD0070 | [18] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0226 | AC11 | HEK-293T | C214(1.12) | LDD1509 | [19] |
| LDCM0270 | AC15 | HEK-293T | C214(0.71) | LDD1513 | [19] |
| LDCM0278 | AC19 | HEK-293T | C214(0.79) | LDD1517 | [19] |
| LDCM0283 | AC23 | HEK-293T | C214(0.90) | LDD1522 | [19] |
| LDCM0287 | AC27 | HEK-293T | C214(0.87) | LDD1526 | [19] |
| LDCM0290 | AC3 | HEK-293T | C214(1.00) | LDD1529 | [19] |
| LDCM0292 | AC31 | HEK-293T | C214(0.75) | LDD1531 | [19] |
| LDCM0296 | AC35 | HEK-293T | C214(0.85) | LDD1535 | [19] |
| LDCM0300 | AC39 | HEK-293T | C214(0.81) | LDD1539 | [19] |
| LDCM0305 | AC43 | HEK-293T | C214(0.88) | LDD1544 | [19] |
| LDCM0309 | AC47 | HEK-293T | C214(1.08) | LDD1548 | [19] |
| LDCM0314 | AC51 | HEK-293T | C214(1.15) | LDD1553 | [19] |
| LDCM0318 | AC55 | HEK-293T | C214(0.79) | LDD1557 | [19] |
| LDCM0322 | AC59 | HEK-293T | C214(1.13) | LDD1561 | [19] |
| LDCM0327 | AC63 | HEK-293T | C214(0.73) | LDD1566 | [19] |
| LDCM0334 | AC7 | HEK-293T | C214(0.99) | LDD1568 | [19] |
| LDCM0156 | Aniline | NCI-H1299 | 11.93 | LDD0403 | [1] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [14] |
| LDCM0379 | CL11 | HEK-293T | C214(1.26) | LDD1583 | [19] |
| LDCM0406 | CL19 | HEK-293T | C214(1.07) | LDD1610 | [19] |
| LDCM0411 | CL23 | HEK-293T | C214(1.23) | LDD1615 | [19] |
| LDCM0420 | CL31 | HEK-293T | C214(0.99) | LDD1624 | [19] |
| LDCM0424 | CL35 | HEK-293T | C214(1.79) | LDD1628 | [19] |
| LDCM0433 | CL43 | HEK-293T | C214(1.44) | LDD1637 | [19] |
| LDCM0437 | CL47 | HEK-293T | C214(1.78) | LDD1641 | [19] |
| LDCM0446 | CL55 | HEK-293T | C214(1.22) | LDD1649 | [19] |
| LDCM0450 | CL59 | HEK-293T | C214(1.96) | LDD1653 | [19] |
| LDCM0459 | CL67 | HEK-293T | C214(1.05) | LDD1662 | [19] |
| LDCM0462 | CL7 | HEK-293T | C214(0.93) | LDD1665 | [19] |
| LDCM0464 | CL71 | HEK-293T | C214(1.40) | LDD1667 | [19] |
| LDCM0472 | CL79 | HEK-293T | C214(0.95) | LDD1675 | [19] |
| LDCM0477 | CL83 | HEK-293T | C214(1.47) | LDD1680 | [19] |
| LDCM0486 | CL91 | HEK-293T | C214(1.09) | LDD1689 | [19] |
| LDCM0490 | CL95 | HEK-293T | C214(1.14) | LDD1693 | [19] |
| LDCM0028 | Dobutamine | HEK-293T | 6.27 | LDD0180 | [5] |
| LDCM0120 | HHS-0701 | DM93 | Y179(13.74) | LDD0268 | [6] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [14] |
| LDCM0022 | KB02 | A431 | C214(1.55) | LDD2258 | [3] |
| LDCM0023 | KB03 | A431 | C214(1.34) | LDD2675 | [3] |
| LDCM0024 | KB05 | RPMI-7951 | C214(1.48) | LDD3410 | [3] |
| LDCM0030 | Luteolin | HEK-293T | 11.44 | LDD0182 | [5] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0233 | [14] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C214(1.00) | LDD2107 | [4] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C214(0.58) | LDD2133 | [4] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Alpha-synuclein (SNCA) | Synuclein family | P37840 | |||
| Wolframin (WFS1) | . | O76024 | |||
Other
References
